Greg Martin

How do bacteria infect plants and how do plants defend themselves from such attacks?
The long-term goal of research in the Martin laboratory is to use knowledge gained about the molecular basis of plant-pathogen interactions to develop plants with increased natural resistance to diseases. Such plants would require fewer applications of pesticides producing economic and environmental benefits while providing food for consumers with less pesticide residue.
The Martin laboratory studies the molecular basis of bacterial infection processes and the plant immune system. The research focuses on speck disease which is caused by the infection of tomato leaves with the bacterial pathogen Pseudomonas syringae pv. tomato. This is an economically important disease that can decrease both the yield and quality of tomato fruits. It also serves as an excellent experimental system for studying the molecular mechanisms that underlie plant-pathogen interactions and how they have evolved. Many experimental resources, including an increasing number of genome sequences, are available for both tomato and P. s. pv. tomato. Current work relies on diverse experimental approaches involving methods derived from the fields of biochemistry, bioinformatics, cell biology, forward and reverse genetics, genomics, molecular biology, plant breeding, plant pathology and structural biology.
Research in the Martin lab is supported, in part, by: USDA-BARD IS-4931-16C, NSF IOS-1451754 and NSF-IOS-1546625
See more...
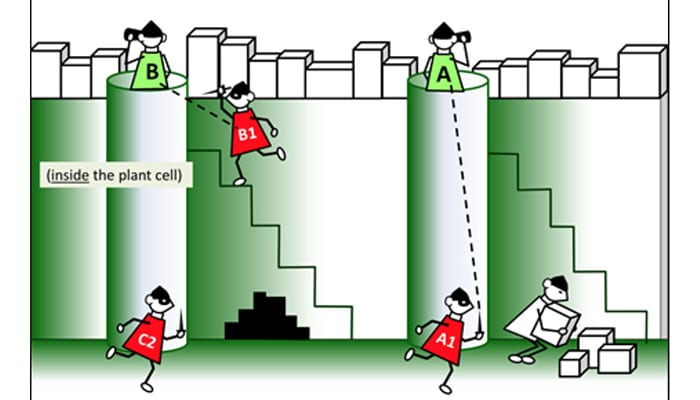
In the interaction of Pseudomonas with tomato, the plant responds rapidly to a potential infection by detecting certain conserved molecules expressed by the pathogen. At this stage, the pathogen uses a specialized secretion system to deliver virulence proteins, such as AvrPto and AvrPtoB, into the plant cell. These pathogen proteins suppress early host defenses and thereby promote disease susceptibility. Some tomato varieties express a resistance gene, Pto, which encodes a protein that detects the presence of AvrPto or AvrPtoB and activates a second strong immune system that halts the progression of bacterial speck disease.
The Martin lab is currently studying many aspects of the molecular mechanisms that underlie the bacterial infection process and the plant response to infection. One project takes advantage of the genetic natural variation present in wild relatives of tomato to identify new genes that contribute to plant immunity. These genes provide insights into the plant immune system and also can be bred into new tomato varieties to enhance disease resistance. A second project relies on next-generation sequencing methods to identify tomato genes whose expression increases during the interaction with P. s. pv. tomato. The expression of these genes is then reduced by using virus-induced gene silencing or the genes are mutated using CRISPR/Cas9 to test whether they make a demonstrable contribution to immunity.
Links
.
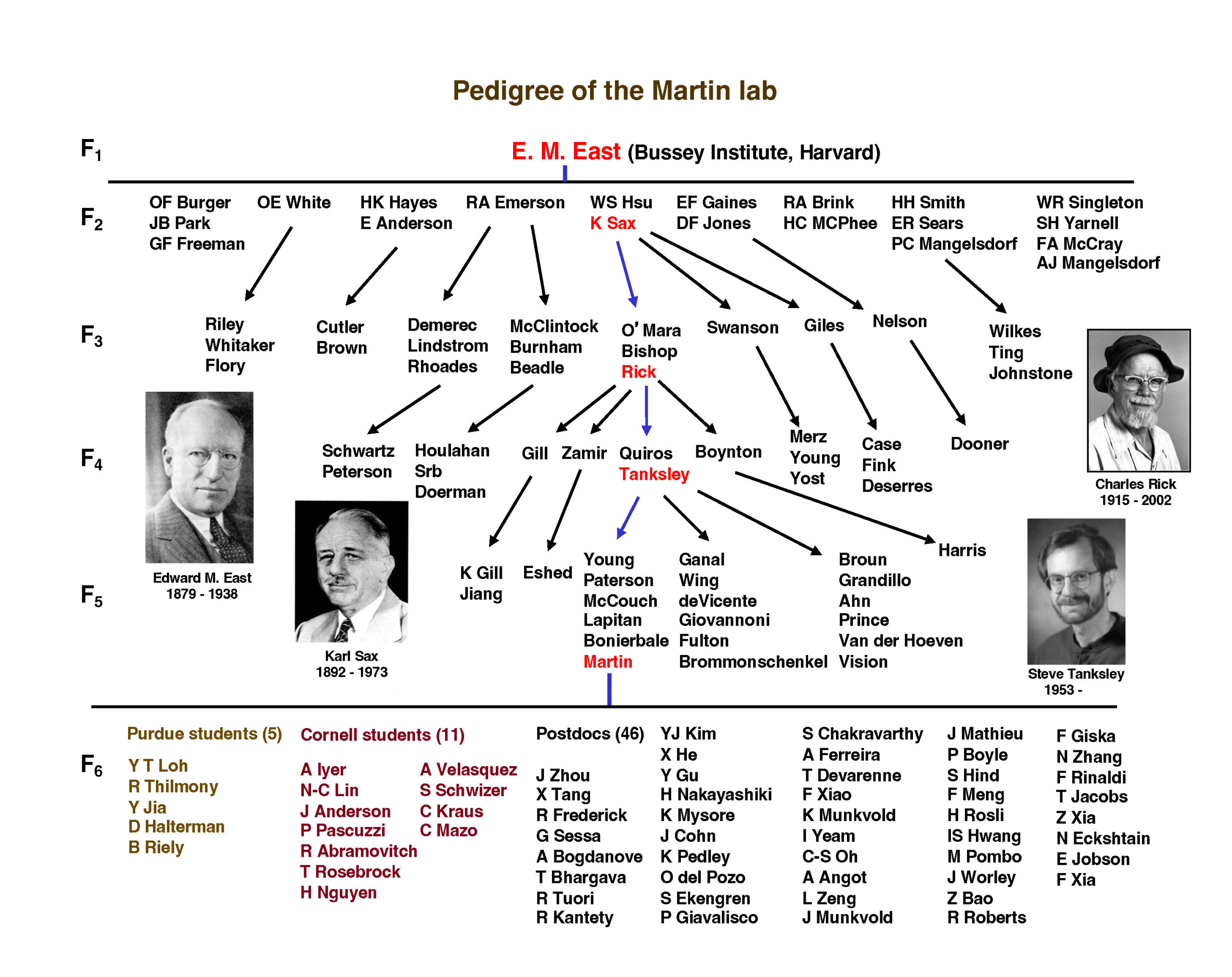
Martin Lab Pedigree
Evolution of immunity
Evolution of pathogen recognition and defense responses in wild species of tomato and tomato heirloom varieties.
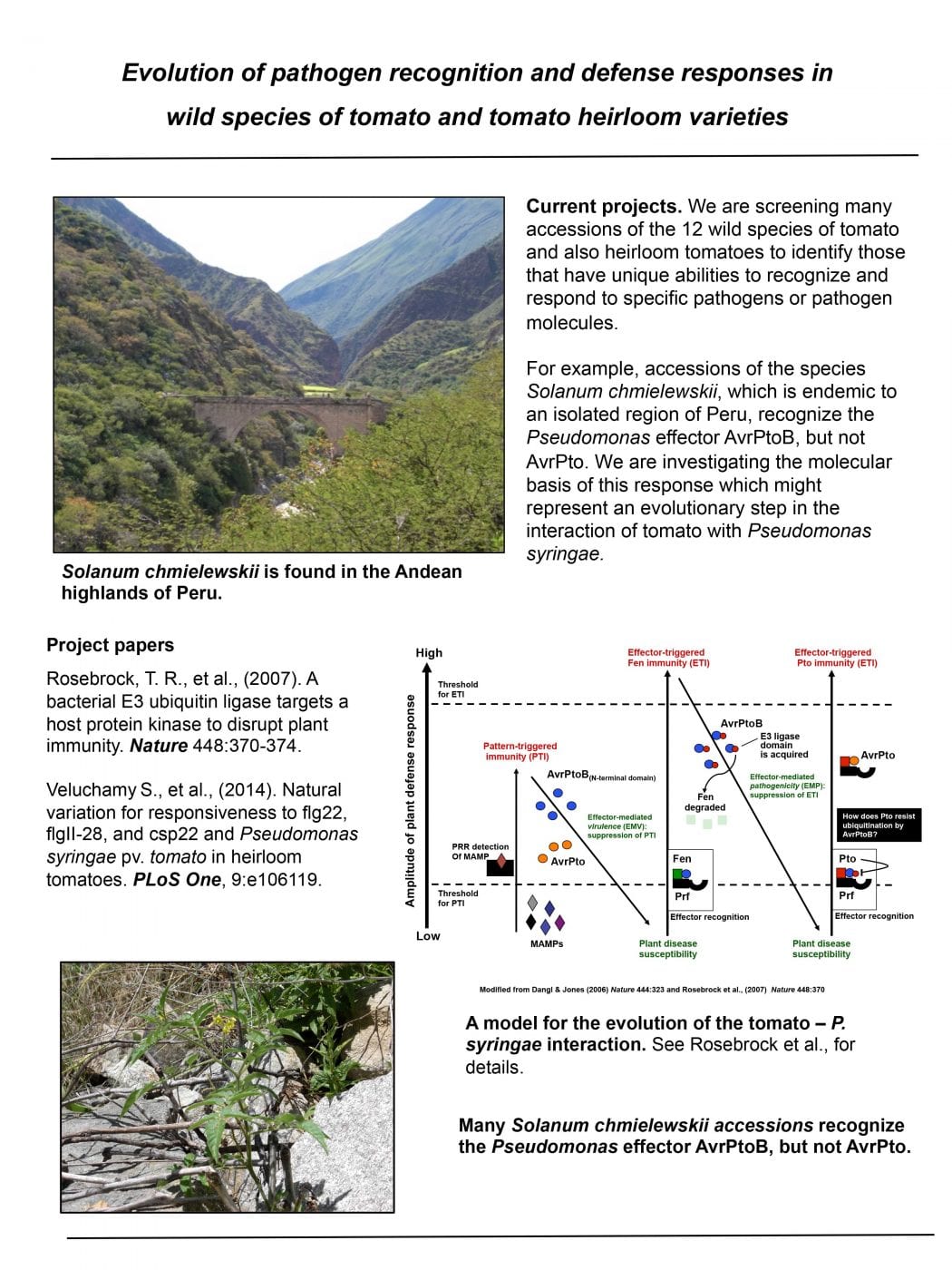
Pattern-triggered immunity
The plant immune system 1: Mechanisms underlying recognition and response to microbe-associated molecular patterns.
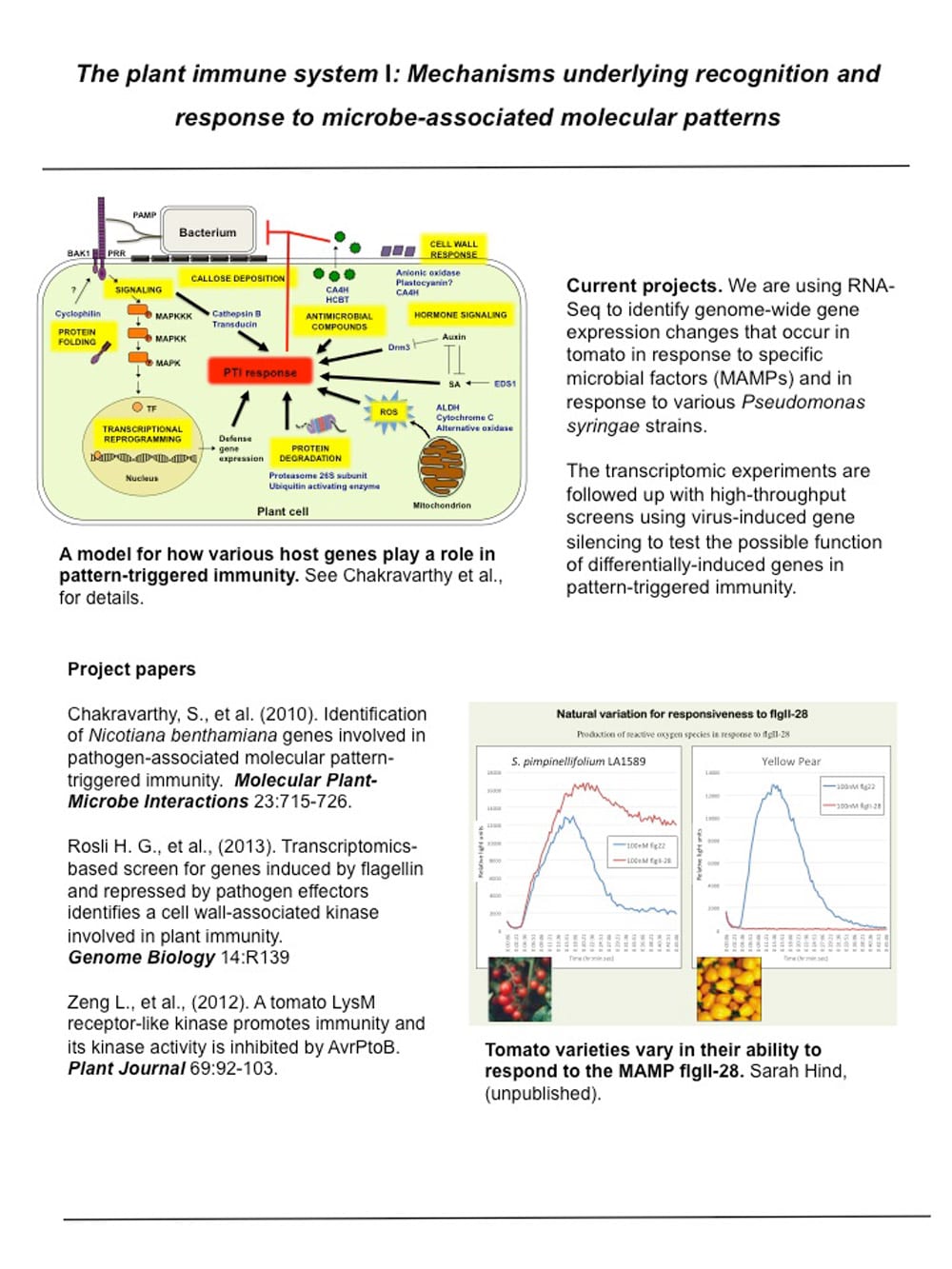
Effector-triggered immunity
The plant immune system II: Mechanisms underlying recognition and response to pathogen effector proteins.
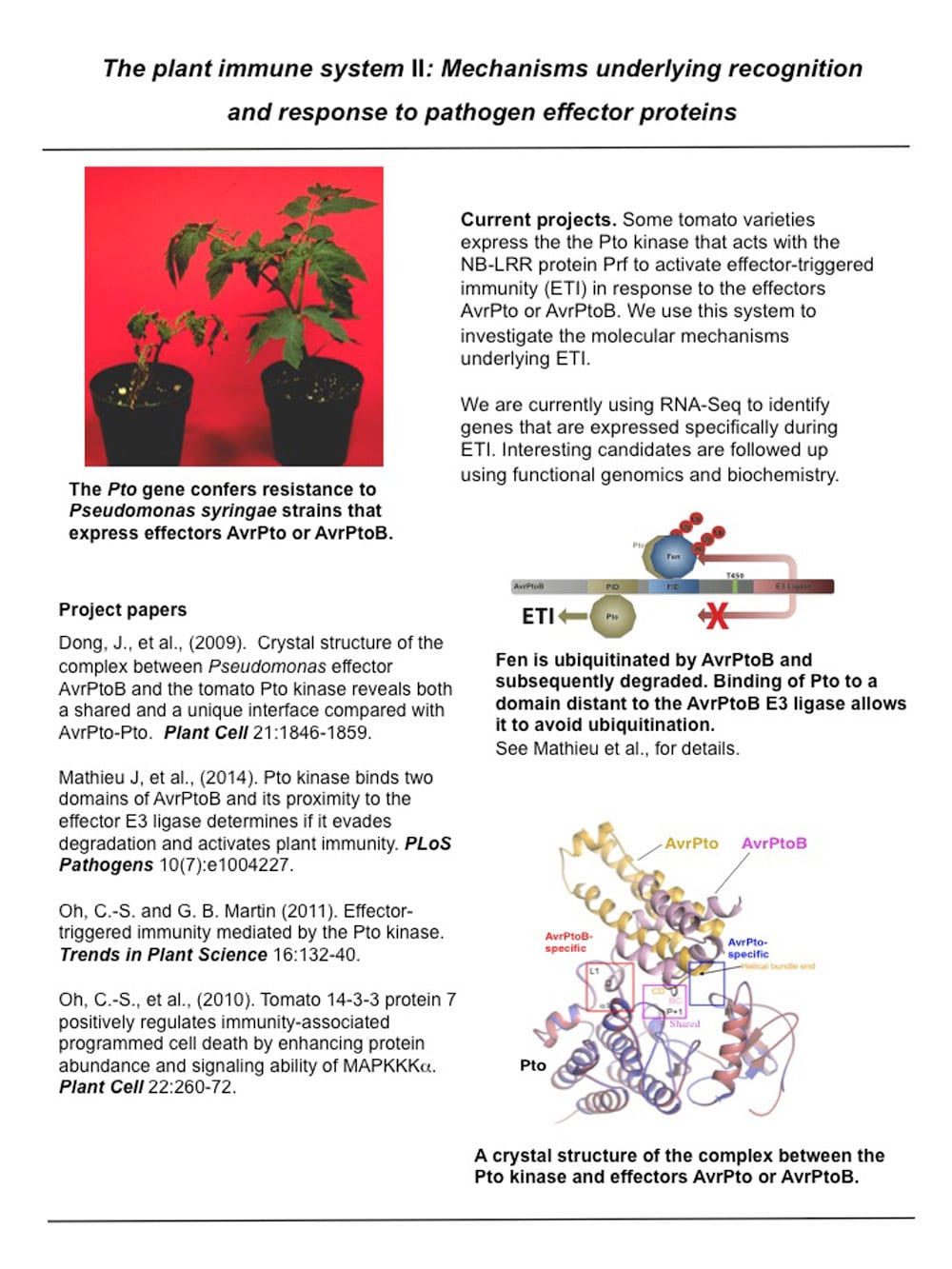
AvrPto virulence mechanisms
Virtulence mechanisms of the AvrPto effector protein from Pseudomonas syringae pv. tomato.
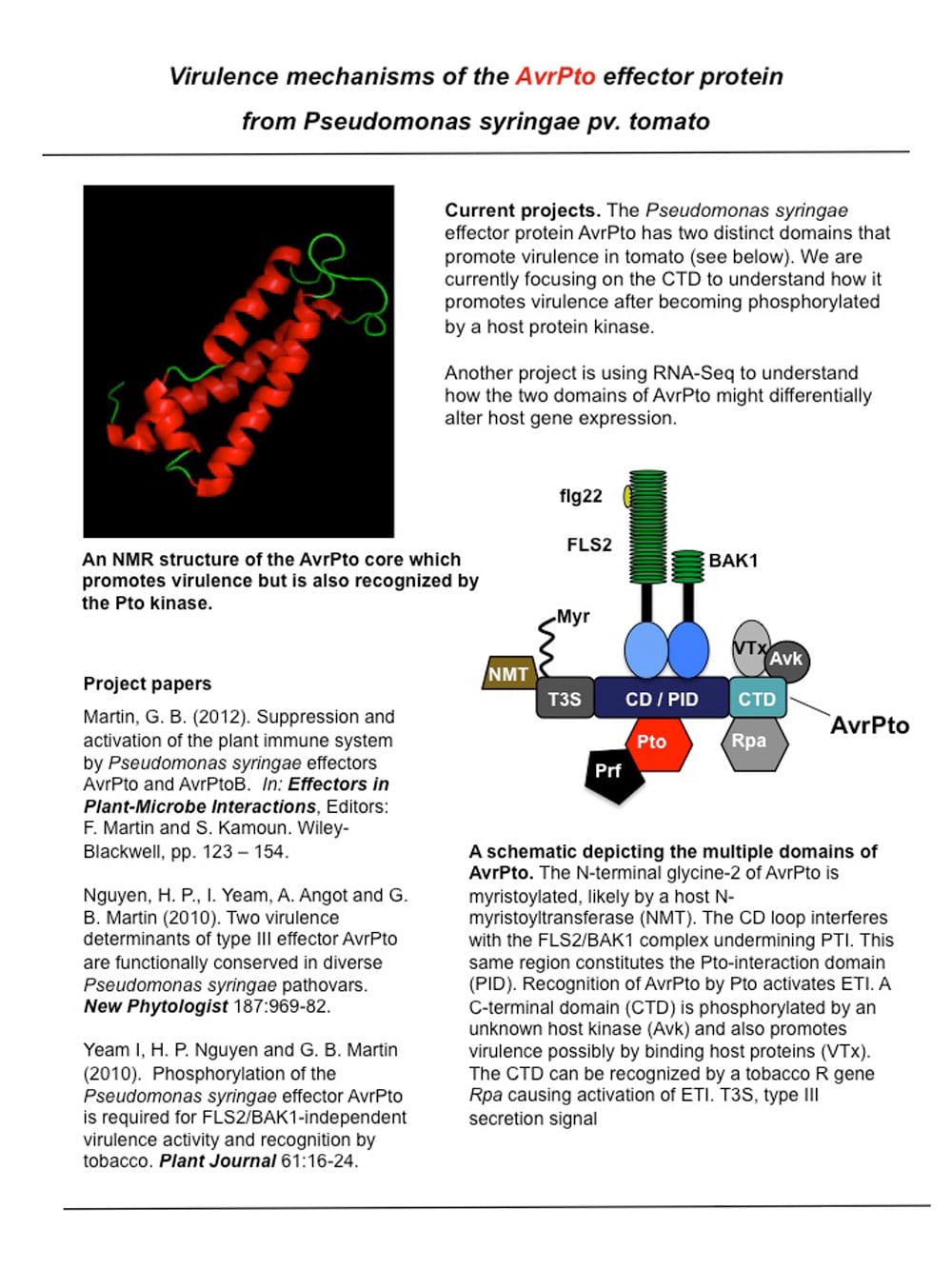
Methods to study plant immunity
Development of methods and resources to study the plant immune system.
Nicotiana benthamiana genome project
Nicotiana benthamiana genome project.

-
Interstellar: Interview with Dr. Greg Martin, newly elected member of the National Academy of Sciences
Interview of Dr. Martin conducted by Haesong Kim, PhD student at Pohang University of Science and Technology, Korea. Read more » -
Wild tomato genome will benefit domesticated cousins
Wild relatives of crops are becoming increasingly valuable to plant researchers and breeders. During the process of domestication, crops tend to lose many genes, but wild relatives often retain genes […] Read more » -
BTI Welcomes Summer Student Interns
On May 31, Boyce Thompson Institute welcomed 41 of the country’s brightest undergraduate students from universities around the country to experience the life of a researcher for 10 weeks. Ten […] Read more »
-
BTI’s Greg Martin Elected to National Academy of Sciences
Greg Martin, Boyce Schulze Downey Professor at the Boyce Thompson Institute and Professor in the School of Integrative Plant Science (SIPS) at Cornell University, has been elected to the National […] Read more » -
Tomato’s Wild Ancestor Is a Genomic Reservoir for Plant Breeders
Thousands of years ago, people in the region now known as South America began domesticating Solanum pimpinellifolium, a weedy plant with small, intensely flavored fruit. Over time, the plant evolved […] Read more » -
Newly Identified Gene Grants Tomatoes Resistance to Bacterial Speck Disease
Bacterial speck disease, which reduces both fruit yield and quality, has been a growing problem in tomatoes over the last five years. Because the culpable bacterium, Pseudomonas syringae, prefers a […] Read more » -
Congratulations Spring 2020 Graduates!
We are pleased to announce that six BTI researchers received their degrees from Cornell University this spring. Congratulations to our newest alumni: Jason Hoki, Schroeder lab, PhD in Chemistry & […] Read more » -
Cluster Hire Yields Three New Faculty Members
Boyce Thompson Institute is pleased to announce the hiring of three faculty members as part of its new and innovative “cluster hire” approach. Out of 113 applicants, the three people […] Read more » -
Congratulations to BTI’s PhD Graduates!
We are pleased to announce that seven Boyce Thompson Institute researchers received their PhD degrees during the Cornell University commencement ceremony on May 26. Congratulations to our newest alumni: Mariko […] Read more » -
Inaugural BTI Alumni Recognition Awards
It is with great enthusiasm and pride that Boyce Thompson Institute (BTI) will recognize the first recipients of BTI’s Alumni Recognition Awards during the 2019 PGS Career Symposium on April […] Read more » -
New CRISPR database to catalyze collaborations
Recently developed gene editing tools like CRISPR/Cas enable scientists to figure out the functions of myriad plant genes. While these studies could eventually lead to the creation of crops with […] Read more » -
Hot tomatoes! MPMI Cover features BTI research
This month, the cover of Molecular Plant-Microbe Interactions features a publication by Simon Schwizer from the Martin Lab at BTI that furthers our understanding of how tomatoes are able to resist infection by Pseudomonas syringae, the causal agent of bacterial speck, a common disease in upstate NY. Read more » -
A New Method for Studying Plant-Germ Warfare
Researchers in the Martin lab develop a new technique to study the arms race between plants and the bacteria that infect them. Read more » -
Collaboration to Identify New Sources of Disease Resistance in Tomato
Professor Greg Martin and colleagues received an NSF grant to pursue research into resistance against bacterial speck disease in tomatoes. Read more » -
SolGenomics Meeting Has Newest Advances in Nightshades
Many BTI researchers will present their latest research at the 13th annual SolGenomics Conference, Sept. 12-16 in Davis, California. Read more » -
New System in Tomato’s Defense against Bacterial Speck Disease
Researchers at BTI and Virginia Tech find a new bacterial detector in tomatoes that could help other crop plants to be more disease resistant. Read more » -
BTI Faculty Honor Former Advisor, Steve Tanksley
Tanksley made invaluable contributions to plant breeding and genetics, laying the foundation for targeted crop improvements to increase food security. Read more » -
BTI Intern Picks Up Awards on His Way to Berkeley
Patel is about to graduate from Cornell--and the Martin lab--to return to his native California to continue his career in plant science. Read more » -
High-throughput CRISPR Vector Construction Using Tomato Hairy Roots
In a JOVE publication, postdoctoral researcher Thomas Jacobs uses tomato hairy roots to demonstrate how multiple CRISPR vectors can be constructed in parallel in a single cloning reaction. Read more » -
The “Speck”-ter Haunting New York Tomato Fields
The BTI tomato field experienced a damaging outbreak of bacterial speck disease this summer, but BTI's Greg Martin has identified genetic regions in a wild tomato species that may make future varieties immune to these devastating bacterial strains. Read more » -
The Perks of Being a Mentor
Summer internships at BTI let students try on the life of a scientist for a few months, while attending a variety of talks, trainings and social events. But what’s in it for the mentors, who painstakingly train them? Read more » -
Scientists Find New Tool for Pathogen to Pillage Plants
“The more we understand the molecular mechanisms involved in the infection process and in plant resistance, the more effective we’re going to be in breeding resistant crops,” said Professor Greg Martin. Read more » -
BTI Hosts Flash Science! Speaking Competition
BTI Professor Emeritus Robert Kohut initiates competition at BTI to give early-career scientists an opportunity to communicate with the general public and practice their “elevator speech.” Read more » -
New Website Is Portal for Nicotiana benthamiana Experimental Resources
This Nicotiana benthamiana web site shares papers, results, tools, protocols, and other materials from researchers using NB as a study plant. Read more » -
A Wild Relative of Tobacco Offers Insight into Molecular Plant Biology at BTI
The genome of an experimentally important relative of tobacco has been sequenced by US and Canadian researchers. Read more » -
Draft Genome Sequence for Nicotiana benthamiana Released in 2012
Scientists at the Boyce Thompson Institute for Plant Research (BTI) released a draft sequence of the Nicotiana benthamiana genome which is accessible through the SGN BLAST tool and can be […] Read more » -
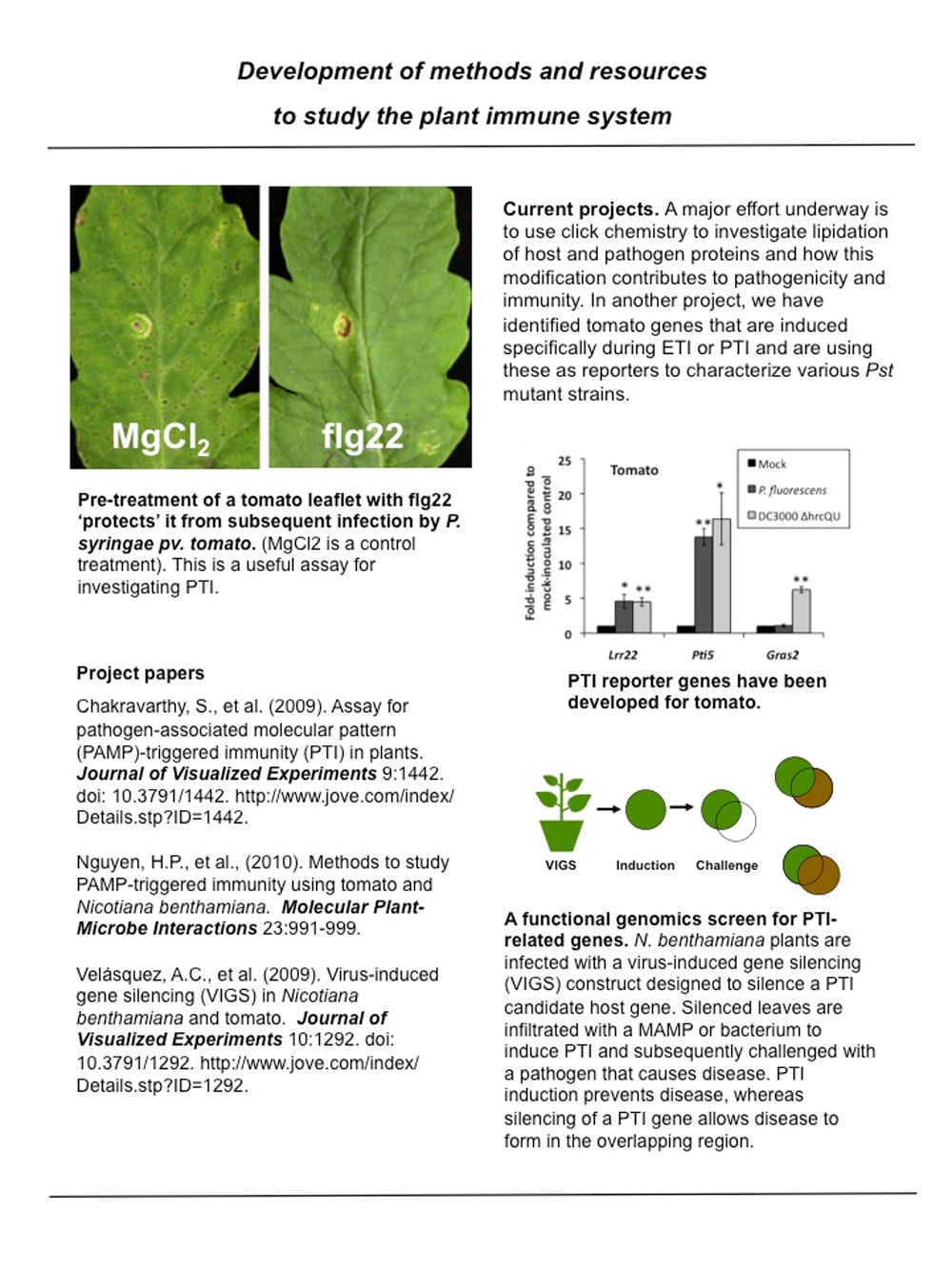
Assay for PAMP-Triggered Immunity in Plants
Research associate Suma Chakravarthy from Greg Martin’s lab, shows the procedure Assay for Pathogen-Associated Molecular Pattern (PAMP)-Triggered Immunity (PTI) in Plants in a JOVE publication. Read more » -
VIGS in Nicotiana benthamiana and Tomato
Graduate student André Velásquez from Greg Martin’s lab shows how to do Virus-induced Gene Silencing (VIGS) in Nicotiana benthamiana and Tomato in a JOVE publication. Read more » -
Tracy Rosebrock Interviewed by Nature Magazine
Tracy Rosebrock, a former BTI graduate student in the laboratory of Greg Martin, discusses how some bacteria suppress plant immunity. Read more »
Protocols
The Martin laboratory studies the molecular basis of plant immunity and bacterial pathogenesis. Our focus is on the infection of tomato by Pseudomonas syringae pv. tomato as this interaction results in bacterial speck, an economically important disease, and also serves as a powerful model system for understanding fundamental mechanisms involved in plant-pathogen biology. On the bacterial side, we study virulence proteins and associated mechanisms that the pathogen uses to interfere with the plant immune response. On the plant side, we identify and characterize genes, proteins and molecular mechanisms that play a role in host immunity and susceptibility. Our work relies on natural variation for these traits that is present in cultivated tomato and in the 12 wild relatives of tomato that originated in South America. For the characterization of both plant and bacterial genes and proteins, we use a variety of experimental approaches including biochemistry, bioinformatics, genetics, genomics, molecular biology, and structural biology.
Examples of research projects in my laboratory include: 1) Using tomato varieties that have natural variation in their immune system to clone and characterize the genes responsible; 2) Using CRISPR/Cas9 genome-editing methods to mutate immunity-associated genes and investigate alterations in the plant defense system; and 3) Investigating bacterial proteins that play a key role in promoting pathogenesis and virulence.
Representative publication: https://nph.onlinelibrary.wiley.com/doi/abs/10.1111/nph.15788
Interns in the Martin Lab are funded by NSF IOS-1451754 and REU-1358843.
Internship Program | Projects & Faculty | Apply for an Internship