Magdalena Julkowska


Plants adjust their development to environmental conditions. In Jukowska lab we are interested in how the environment shapes plant architecture, and what plant architectural traits contribute to overall environmental resilience. In the past, we explored Root System Architecture with simple descriptive models, like Root-Fit (Julkowska et al., 2014), as well as identified underlying genes using forward genetics (Julkowska et al., 2017). We also looked at salt stress-induced changes in root-to-shoot ratio and identified a gene of unknown function (DUF247) to play a significant role in salt stress tolerance.
The current focus of the Julkowska lab is to explore stress-induced changes in plant architecture across stress-tolerant species, including wild tomato (Solanum pimpinellifolium), cowpea (Vigna unguiculata) and tepary bean (Phaseolus acutifolius). In collaboration with Dr Chandrasekhar and Dr Lobet, we are working on new ways to describe plant architectures, by incorporating Pareto front optimality calculations (Chandrasekhar & Julkowska, 2022). Moreover, in collaboration with Dr Andrew Nelson’s lab (BTI), we are working on developing widely accessible plant phenotyping setups using Raspberry Pi and Arduino microcontrollers, and data processing pipelines based on PlantCV and R (Yu & Sussman et al., 2023).
Diversity, Equity and Inclusion Statement
We recognize that academia is suffering from low diversity, equity and inclusion, and we recognize that science operates within the context of wider societal, economic and political issues. We appreciate everyone’s diverse perspectives and strive to create a safe, healthy and inclusive lab environment, that allows everyone to reach their full potential, work towards their own goals, and promote the professional development of lab members at all career stages. Our lab practices to ensure an equitable, inclusive and diverse environment include:
- Frequent mentoring sessions (for the first 3 months – once per month, afterwards every 6 months), weekly one-on-one meetings with Magda, weekly group meetings, and recognizing that clarity in instructions and expectations is kindness.
- Providing and receiving constructive feedback both ways on scientific performance, career goals, professional development and what mentoring style works for each lab member
- Normalizing and learning from failure
- Providing opportunities for science communication and participation in the scientific community through peer review, conference attendance and representation of the lab in institutional presentations and outreach activities at all career stages
- Paid undergraduate internships and overtime for non-exempt employees
- Recognizing all colleagues to contain a multitude of identities, obligations and passions outside their work.
Please contact Magda for the most recent version of the lab handbook, to gain further insight into lab culture.

Links
Please visit the BTI Plant Phenotyping Facility webpage!
Past Lab Members
 |
 |
 |
|---|---|---|
Drought And Salt Stress-Induced Changes In Tepary Bean (Supported by USDA-NIFA Seed Grant)

 Tepary bean (Phaseolus acutifolius) is a native crop grown in arid and semi-arid regions of the Southwestern U.S., Northwestern Mexico, and Central America. While genetic and genomic resources have been developed for tepary bean breeding, we are working to improve the phenomic and genomic resources for tepary beans to accelerate breeding and discovery of genetic components associated with improved agronomic traits and environmental resilience.
Tepary bean (Phaseolus acutifolius) is a native crop grown in arid and semi-arid regions of the Southwestern U.S., Northwestern Mexico, and Central America. While genetic and genomic resources have been developed for tepary bean breeding, we are working to improve the phenomic and genomic resources for tepary beans to accelerate breeding and discovery of genetic components associated with improved agronomic traits and environmental resilience.
We isolated DNA from 400 tepary bean accessions, and genome sequencing data was used for SNP calling, in collaboration with Dr Andrew Nelson, Dr Timothy Porch (USDA-ARS) and Dr Duke Pauli (Uni. of Arizona). The entire collection was also screened for drought-induced changes in the shoot growth and architecture using the Raspberry Pi PhenoCage setup, and salt-induced changes in the root growth and architecture using the paperponics setup developed in Dr Miguel Pineros’ lab (USDA-ARS). The results of this project will be used for establishing Genome-Wide Association Study pipelines, and identifying genetic components underlying plant architecture, and stress resilience
Fundamental Understanding of Stress-resilience in Cowpea (Supported by Triad funding)
 Cowpea (Vigna unguiculata) is a legume crop native to West Africa, resilient to many biotic and abiotic factors. Hayley Sussman and Olga Khmelnitsky developed protocols for screening cowpea’s responses to salt and drought stress for soil-grown plants. Hayley Sussman screened a global natural diversity panel, consisting of 368 accessions, for their responses to drought, and is currently preparing the data for the Genome-Wide Association Study, to identify genetic components underlying drought stress tolerance
Cowpea (Vigna unguiculata) is a legume crop native to West Africa, resilient to many biotic and abiotic factors. Hayley Sussman and Olga Khmelnitsky developed protocols for screening cowpea’s responses to salt and drought stress for soil-grown plants. Hayley Sussman screened a global natural diversity panel, consisting of 368 accessions, for their responses to drought, and is currently preparing the data for the Genome-Wide Association Study, to identify genetic components underlying drought stress tolerance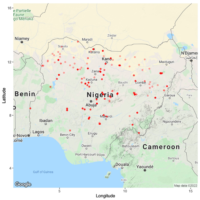 .
.
Moreover, Hayley Sussman is also working on collecting material for DNA isolation and sequencing of the Nigerian cowpea diversity panel, which was shared with us by Dr Ramatu Aliyu (Ahmadu Bello University). By developing the Nigerian cowpea panel, in collaboration with Dr Aliyu, Dr Andrew Nelson and Dr Suzy Stickler (BTI), we aim to increase the number of identified SNPs, and develop pipelines for cowpea Genome-Wide Association Studies / RNA-sequencing, allowing quick trait evaluation and associations with genetic markers. This project, funded by the Triad foundation, will significantly contribute to future breeding efforts and accelerate the fundamental understanding of environmental resilience.
Salt-Induced Changes In Root Architecture Of Wild Tomato (S. pimpinellifolium)
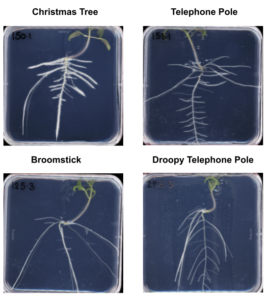
Solanum pimpinellifolium is the closest relative to cultivated tomato, and it is native to the west coast of South America. Different accessions of S. pimpinellifolium were collected from a wide variety of habitats and showed high diversity in their salt stress response in root architecture and salinity-induced changes therein. We recognized four visually distinct topologies of root architecture, that can be distinguished using Pareto Front optimality measures. We are working to expand the Pareto front calculations to improve the physiological representation of the constraints to root system architecture in the current model in collaboration with Dr Arjun Chandrasekhar (Southwestern University). Currently, Dr Maryam Rahmati-Ishka is investigating how we can alter distinct topologies using hormonal treatments, and what are the consequences of these topologies for sodium ion accumulation. Dr Rahmati-Ishka is also investigating the genetic components underlying salt stress sensitivity of lateral root development, by crossing two tomato lines with contrasting salt stress responses. Dr Julkowska is performing GWAS on collected root architecture data to identify candidate genes underlying root development and salt stress-induced changes. All the work in S. pimpinellifolium is currently focusing on salt stress. Hayley Sussman is responsible for the propagation of S. pimpinellifolium collection.
How does Salt Stress Affect Root-To-Shoot Ratio in Arabidopsis?
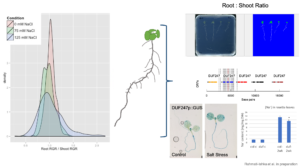 Salt stress rapidly impacts the growth and development of different plant organs to different extents, leading to alterations in plant development. Alterations to whole plant architecture, such as root-to-shoot ratio, have not been extensively explored thus far. Together with KAUST Visualization Lab, we developed a tool that quantifies green and white pixels, reflecting changes in root and shoot surface in Arabidopsis seedlings. Through forward genetics, we identified genetic components underlying salt stress-induced changes in root-to-shoot ratio including DUF247, encoding Domain of Unknown Function. Currently, Dr Rahmati-Ishka is examining the loss-of-function mutants of DUF247 using plate assays and soil-grown plants, as well as the subcellular localization of fluorophore-tagged DUF247 and GUS promoter fusion lines. Since the identified DUF247 is located in the genetic region containing other DUF247 genes, Hayley Sussman investigated the available loss-of-function mutants of the DUF247 genes located on Arabidopsis chromosome 3. Hayley Sussman generated transgenic lines targeting multiple DUF247 genes, as well as over-expression lines of specific DUF247 domains, that are currently being selected for homozygosity. Our preliminary results suggest that identified DUF247 is a negative regulator of Casparian strip formation and limits potential salt stress tolerance.
Salt stress rapidly impacts the growth and development of different plant organs to different extents, leading to alterations in plant development. Alterations to whole plant architecture, such as root-to-shoot ratio, have not been extensively explored thus far. Together with KAUST Visualization Lab, we developed a tool that quantifies green and white pixels, reflecting changes in root and shoot surface in Arabidopsis seedlings. Through forward genetics, we identified genetic components underlying salt stress-induced changes in root-to-shoot ratio including DUF247, encoding Domain of Unknown Function. Currently, Dr Rahmati-Ishka is examining the loss-of-function mutants of DUF247 using plate assays and soil-grown plants, as well as the subcellular localization of fluorophore-tagged DUF247 and GUS promoter fusion lines. Since the identified DUF247 is located in the genetic region containing other DUF247 genes, Hayley Sussman investigated the available loss-of-function mutants of the DUF247 genes located on Arabidopsis chromosome 3. Hayley Sussman generated transgenic lines targeting multiple DUF247 genes, as well as over-expression lines of specific DUF247 domains, that are currently being selected for homozygosity. Our preliminary results suggest that identified DUF247 is a negative regulator of Casparian strip formation and limits potential salt stress tolerance.
Molecular mechanisms underlying reduced lateral root development in UAS-HKT1 lines
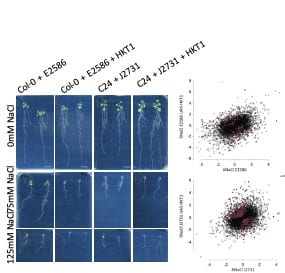 We discovered that high expression of Arabidopsis High-Affinity potassium (K+) Transporter (AtHKT1-1), sequestering sodium from the transpiration stream in root pericycle, results in increased salinity tolerance (Julkowska et al., 2017). Pericycle-specific over-expression of HKT1 resulted in reduced lateral root development under saline conditions. Interestingly, this effect seems to be specific to dicotyledonous species, as wheat lines with high HKT1 expression do not show a reduction in secondary lateral roots. Dr Julkowska examined the transcriptional changes underlying altered lateral root development downstream of HKT1 through comparative transcriptomic analysis of two lines with stelar over-expression of HKT1. Among the transcripts that were expressed differentially between UAS-HKT1 over-expression lines and their background lines, Dr Julkowska found genes involved in cell differentiation, ABA signalling and ion transport. Further validation of identified candidate genes using CRISPR-Cas9 knockouts is being currently being performed by Dr Julkowska, while over-expression lines of the genes are generated in Dr Mark Tester’s lab (KAUST). Further studies of generated mutant lines will enhance our understanding of processes controlling plant development under abiotic stress conditions and the importance of root architecture for salinity tolerance.
We discovered that high expression of Arabidopsis High-Affinity potassium (K+) Transporter (AtHKT1-1), sequestering sodium from the transpiration stream in root pericycle, results in increased salinity tolerance (Julkowska et al., 2017). Pericycle-specific over-expression of HKT1 resulted in reduced lateral root development under saline conditions. Interestingly, this effect seems to be specific to dicotyledonous species, as wheat lines with high HKT1 expression do not show a reduction in secondary lateral roots. Dr Julkowska examined the transcriptional changes underlying altered lateral root development downstream of HKT1 through comparative transcriptomic analysis of two lines with stelar over-expression of HKT1. Among the transcripts that were expressed differentially between UAS-HKT1 over-expression lines and their background lines, Dr Julkowska found genes involved in cell differentiation, ABA signalling and ion transport. Further validation of identified candidate genes using CRISPR-Cas9 knockouts is being currently being performed by Dr Julkowska, while over-expression lines of the genes are generated in Dr Mark Tester’s lab (KAUST). Further studies of generated mutant lines will enhance our understanding of processes controlling plant development under abiotic stress conditions and the importance of root architecture for salinity tolerance.
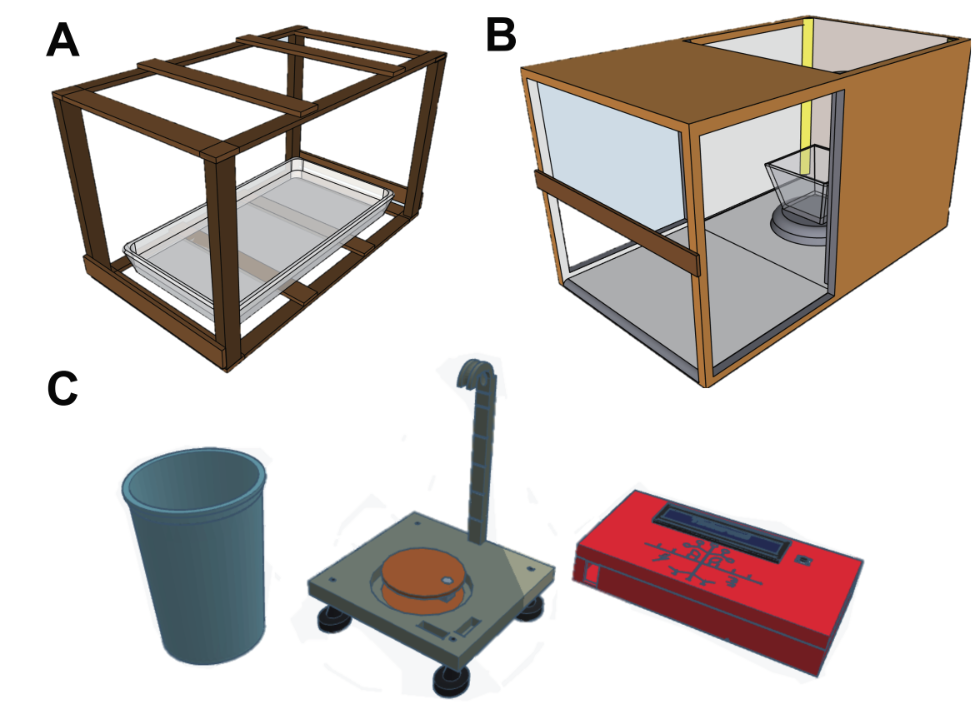
Development of phenotyping tools and protocols (supported by Goelet foundation)
 Studying the drought resilience using a non-destructive experimental setup poses significant opportunities. Traditionally, soil-grown plants are evaluated based solely on the above-ground portions due to easier accessibility. Aeroponics technology stands out as a game-changer, as it provides easy access to roots and shoots for imaging, and offers precise control over water supply. In collaboration with Prof. Perrinne Periot (CU Enngineering) we are developing aeroponics system where roots are misted with a nutrient solution. We have successfully mimicked drought conditions, eliciting physiological responses similar to those of drought-stressed plants grown in soil. As aeroponics allows for rapid changes in water availability, it will permit differentiation between water conservation and true resilience strategies within tolerant genotypes.
Studying the drought resilience using a non-destructive experimental setup poses significant opportunities. Traditionally, soil-grown plants are evaluated based solely on the above-ground portions due to easier accessibility. Aeroponics technology stands out as a game-changer, as it provides easy access to roots and shoots for imaging, and offers precise control over water supply. In collaboration with Prof. Perrinne Periot (CU Enngineering) we are developing aeroponics system where roots are misted with a nutrient solution. We have successfully mimicked drought conditions, eliciting physiological responses similar to those of drought-stressed plants grown in soil. As aeroponics allows for rapid changes in water availability, it will permit differentiation between water conservation and true resilience strategies within tolerant genotypes.

Other germinating projects
- Understanding metabolic contributions to drought stress resilience in cowpea and other legume species
-
BTI Welcomes Summer Student Interns
On May 31, Boyce Thompson Institute welcomed 41 of the country’s brightest undergraduate students from universities around the country to experience the life of a researcher for 10 weeks. Ten […] Read more »