Spring 2018 BCBC Bioinformatics Course
About the Course
We are living in massive data times, and science is not an exception. New sequencing technologies are filling hard disks with terabytes of information, billion of sequences that need to be analyzed in a proper way. But not only sequence data is growing, gene expression and metabolite concentrations are analyzed by the hundreds or thousands, in a way that makes it difficult, if not impossible, to use familiar tools such as Excel. In this new world, bioinformatic skills are needed, not only by computational biologists, but by biologists and biochemists who find themselves analyzing many genes, proteins or metabolites at the same time.With this perspective, we aim to further bioinformatic skills within the postdoc community at Boyce Thompson Institute through this course. We try to keep it as simple as we can, with just one idea: “Show useful tools to resolve common problems found during the *omics data analysis”. For example, if I have two lists of hundred of genes, how can I combine them and find the common ones, or how can I analyze GO terms for my over-expressed genes, or how can download a chromosome region using Jbrowse or… there are dozens of examples.
Before the Course: Setting Up the BCBC Course Virtual Machine
 6. Select the option File > Import Appliance
6. Select the option File > Import Appliance 7. Click “Open Appliance”. Select the .ova file and click “Continue”.
7. Click “Open Appliance”. Select the .ova file and click “Continue”. 8. Enable “Reinitialize the MAC address of all the network cards” and click “Import”.
8. Enable “Reinitialize the MAC address of all the network cards” and click “Import”.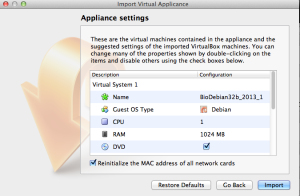 Troubleshooting
Troubleshooting
- I have a Mac computer, I am using Safari and I can not download the whole file.
Safari has some problems to deal with big files and the ftp site, but you can resolve it by using Firefox or Chrome.
- I have downloaded the file and when I tried to use the file it says “File Corrupted” or something similar.
The download was likely interrupted at some point. We recommend using a wire connection to download the file because it is large and the chances that something fail during the download are high. Once you have download the file you can do a md5sum to verify that the file is complete. You can find the md5sum codes at: md5sum. In Mac computer you only need to open the terminal, type “cd ~/Downloads” (Or the dir where you downloaded the file) and then “md5 BCBCBIC2018_debian.ova”. In a Windows computer, you can use WinMD5.
- I have downloaded both the VirtualBox software and the virtual machine file, but it will not run.
Make sure you have a 64-bit machine and have followed the above steps precisely, especially enabling . Come to our office or email us for further troubleshooting if you cannot find the problem.
3/13/18 — UNIX Command-Line Intro, Part 1
Topics covered
- Terminal file system navigation
- Wildcards, shortcuts and special characters
- File permissions
- Compression UNIX commands
- Networking UNIX commands
Estimated Time
- Lecture and exercises: 2:00 h
Materials
3/20/18 — UNIX Command-Line Intro, Part 2
Topics covered
- Basic NGS file formats
- Text files manipulation commands
- Command-line pipelines
- Introduction to bash scripts
Estimated Time
- Lecture and exercises: 2:00 h
Materials
3/27/18 — NGS and RNA-seq
Topics covered
- Background of RNA-seq
- Application of RNA-seq (what RNA-seq can do?)
- Available sequencing platforms and strategy and which one to choose
- RNA-seq data analysis
- Read processing and quality assessment
- De novo assembly
- Alignment to reference genome/transcriptome
- Differentially expressed gene identification
- Downstream analysis using Plant MetGenMAP
Estimated Time
- Lecture and examples: 2:00 h
Materials
4/03/18 — Sequencing, Assembly and Quality Control
Topics covered
- Different sequencing technologies
- Sequence file types and formats
- Genome Assembly
- Annotation
- Quality encoding
- Quality control tools
Estimated Time
- Lecture and examples: 2:00 h
Materials
4/10/18 — Mapping NGS Data
Topics covered
- Overview of NGS sequence assembly
- Reference-guided RNA-seq assembly with HISAT2
- RNA-expression analysis with StringTie
- Overview of other useful tools for NGS analysis
Estimated Time
- Lecture and examples: 2:00 h
Materials
4/17/18 — SNP calling from NGS data
Prerequisites
- The four output .bam files from the previous session “Mapping NGS Data.” Please let us know before class if you missed the previous session, or were unable to complete the exercises, so we can make sure you have the necessary files.
Topics covered
- Overview of SNP calling tools for NGS data
- SNP calling using GATK and Samtools
- SNP annotation and effect prediction with SnpEff
Estimated Time
- Lecture and examples: 2:00 h
Materials
4/24/18 — Introduction to R & Basic R Graphs
Topics covered:
- Brief introduction to R
- Data types
- R graphs
Estimated Time
- Lecture and examples: 2:00 h
Materials
5/01/18 — Differential expression with edgeR
Prerequisites
- Make sure that you have “gene_count_matrix.csv” file in the “Slch04_demo” directory in the Desktop directory of your VM. Please let us know before class if you missed a previous session, or were unable to complete the exercises, and do not have the necessary files.
Topics covered
- General pipeline for differential expression analysis with an emphasis on edgeR
- Data exploration
Estimated Time
- Lecture and examples: 2:00 h
Materials


