As world leaders in plant genome research, Cornell University, Boyce Thompson Institute (BTI), the USDA-ARS, and the U.S. Plant, Soil, and Nutrition Laboratory are host to many outstanding research labs. These research facilities have built on Cornell’s long tradition of research in plant genetics and breeding to develop novel technologies, the application of which has sought to improve the scientific understanding of many aspects of plant biology. The research interests of the labs are quite varied, ranging from identifying disease resistance in crop plants to understanding how plants sense and respond to light. Please click the following text to learn more about the faculty members associated with the research projects in the various summer internship opportunities at BTI.
Internship Projects
Click on the topics below to learn more about available projects and their faculty sponsors. All the projects listed are through our Plant Genome REU program unless otherwise listed in the title. As a reminder, we host two REU programs, Plant Genome REU and Programmable Plant Systems REU. Our Programmable Plant Systems REU program is in collaboration with the Center for Research on Programmable Plant Systems at Cornell University.
For examples of previous projects through our Programmable Plant Systems REU program, please review the list of example projects found here. You will also find interviews with previous Programmable Plant Sytems REU interns who discuss their projects.
Chemical ecology and coevolution of monarchs and milkweeds - Agrawal Lab, Cornell
Project description:
 Our lab studies the ecology and evolution of plant-insect interactions, including aspects of plant defense (the spines and toxins plants produce to reduce herbivory), induced defense (the immune-like system of plants in response to attack), chemical ecology (how interactions between species are mediated by chemistry), and coevolution (long-term reciprocal adaptation and “arms race” evolution when species battle). We work on local biodiversity and especially milkweed plants and monarch butterflies.
Our lab studies the ecology and evolution of plant-insect interactions, including aspects of plant defense (the spines and toxins plants produce to reduce herbivory), induced defense (the immune-like system of plants in response to attack), chemical ecology (how interactions between species are mediated by chemistry), and coevolution (long-term reciprocal adaptation and “arms race” evolution when species battle). We work on local biodiversity and especially milkweed plants and monarch butterflies.
Approaches in our lab are diverse, and typically involve field research, chemical analyses, genetic techniques, and rearing lots of bugs. Our work has advanced both basic questions in ecology, evolution, and plant biology, as well as applications to insect pest management (especially in cucurbit crops) as well as conservation biology (of monarch butterflies).
For more information about Agrawal’s lab, publications, and his blog please click here
Faculty advisor: Anurag Agrawal

Molecular Definition of Plant Organelle Editosomes - Bentolila Lab, Cornell
Project description:
Among post-transcriptional processes that affect gene expression, C-to-U RNA editing in plants exhibits a number of unique features. Editing is confined to the genome-containing plant organelles—chloroplasts and mitochondria. Unlike other types of RNA editing, the purpose of plant RNA editing is not to create multiple proteins from the same transcript—instead, its role is to correct T-to-C mutations in critical locations of transcripts in order to produce functional proteins. A typical vascular plant requires over 600 RNA editing-mediated corrections in its organelle transcripts. A combinatorial approach involving quantitative genetics, biochemistry and genomics has allowed us to identify several components of the editosome, the editing machinery responsible for the modification of C to U on organelle transcripts. In addition to PPR proteins, which act as recognition factors controlling the specificity of the editing reaction, we have identified three other protein families as providing components to the plant editosome. Ultimately the goal of this research is to reconstitute the plant editosome in vitro, and from there pave the way for future genetic engineering of RNAs in plants and other organisms, technology which could benefit the biotechnology industry and society.
Faculty advisor: Stephane Bentolila

TAL effector biology for durable plant disease prevention and DNA targeting applications - Bogdanove Lab, Cornell
Project description:
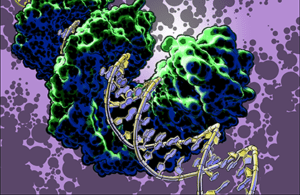
TAL effector PthXo1 bound to its target. Illustration by Jon Bogdanove
Research in the Bogdanove laboratory is directed at understanding mechanisms of bacterial plant pathogenesis and plant defense to develop better means of disease control. We focus in the interactions of plant pathogenic bacteria in the genus Xanthomonas with important crop plants. TAL effectors are injected into plant cells by the bacterium, enter the nucleus, and activate specific plant “susceptibility” (S) genes that contribute to disease development. Immune receptor proteins in some plants recognize incoming TAL effectors and signal a defense response, making the plant resistant to infection. But some strains of the pathogen have evolved variant TAL effectors that can interfere with the function of the immune receptor. Major areas of activity in our lab include 1) identification and characterization of S genes, with an eye toward not only understanding how they work but how we can make them inaccessible to the pathogen; and 2) structural, biochemical, and cell biological studies to understand how TAL effector-specific immune receptors work, with an eye toward engineering them to be impervious to interference by variant TAL effectors. REU opportunities are available in both of these areas.
Faculty advisor: Adam Bogdanove

Virus recognition of insect vectors and collaborations - Casteel Lab, Cornell
Project description:

Numerous studies demonstrate that vector-borne pathogens, such as viruses, influence host characteristics that result in altered host-vector interactions and enhanced virus transmission. We recently demonstrated that viral proteins respond actively to the presence of insect vectors, promoting insect performance and transmission only when needed. We seek to determine the molecular mechanisms that underlie this phenomenon and use this knowledge to develop innovative control strategies using genetic and biochemical approaches. Current focuses are on changes in plant protein turnover and defenses, cell biology, chemical ecology, and protein functions in response to virus infection and insect vectors. Students will receive training in molecular biology, chemistry, genetic engineering, and ecology.
Faculty advisor: Clare Casteel

Bioinformatics and genomics to understand important traits of agricultural crops - Fei Lab, BTI
Using integrated bioinformatics and genomics approaches to understand important traits of agricultural crops.
Project description:
 The development of high-throughput technologies has generated a wealth of system-level information, including genome, transcriptome, proteome, and metabolome data. However, digesting this massive amount of information and applying it in an intelligent and comprehensive manner remains a significant challenge. To address this, Dr. Fei’s group has focused on analyzing and integrating large-scale ‘omics’ datasets to better understand the genetic basis of crop evolution and breeding, as well as the genetic architecture of key agronomic traits.
The development of high-throughput technologies has generated a wealth of system-level information, including genome, transcriptome, proteome, and metabolome data. However, digesting this massive amount of information and applying it in an intelligent and comprehensive manner remains a significant challenge. To address this, Dr. Fei’s group has focused on analyzing and integrating large-scale ‘omics’ datasets to better understand the genetic basis of crop evolution and breeding, as well as the genetic architecture of key agronomic traits.
Faculty advisor: Zhangjun Fei

Quantifying the influence of flower color on plant-pollinator interactions in soybean - Frank Lab, Cornell
Project description:

Xylem-mobile dye moving through a pepper leaf (image credit: Hannah Thomas)
The production of hybrid seeds has been used for over a century to increase crop yields, improve abiotic and biotic stress tolerance, and enhance nutritional quality. Soybean, one of the most important crops in the US, is a self-fertilizing plant with untapped yield and heterotic trait potential. Using a biotechnology approach, the Frank lab is working to transform soybean into an outcrossing crop that can benefit from hybrid breeding. One essential component of this work involves attracting bee pollinators that can promote outcrossing. In this project, students will use plant genetics, video imaging, computational modeling, and microscopy to test the influence of altered floral traits in bee visitation in greenhouse and field settings.
Faculty advisor: Margaret Frank

Carbon-concentrating mechanism in hornworts- Laura Gunn (Plant Biology) and Fay-Wei Li (BTI)
Project Description
Nature’s carbon-fixing enzyme, Rubisco, represents the major point of carbon entry into the biosphere. However, Rubisco is notoriously inefficient, exhibiting a slow catalytic rate and poor discrimination between CO2 and O2. O2-fixation leads to photorespiration, which consumes energy and releases fixed CO2. To enable more efficient photosynthesis, certain organisms have evolved biophysical carbon-concentrating mechanisms (CCM) that partition Rubisco and CO2 into a dedicated subcellular compartment. Pyrenoids are an example of such compartments – organelles comprised of an interconnected matrix of aggregated Rubisco (and associated proteins) that are liquid-liquid phase separated from the stroma (and molecular oxygen). The green alga Chlamydomonas reinhardtii has been the traditional model to study pyrenoid-based CCMs with the hope of installing a similar mechanism into crops to enhance yield. However, hornworts, a lineage of bryophytes, are the only land plants that have a pyrenoid-based CCM. Owing to their much closer relationship to crop plants, lessons learned from hornworts might have higher translational potential than those from Chlamydomonas. We are working to identify and validate putative hornwort pyrenoid components, and determine how they interact with each other in order to develop the blueprints for building a hornwort pyrenoid in crop chloroplasts.
Faculty advisors:

Laura Gunn

Fay-Wei Li
Biogenesis pathway understanding to reimagine CO2 fixation in land plant chloroplasts - Laura Gunn, Cornell
Project Description
Nature’s carbon fixing enzyme, Rubisco, is notoriously inefficient, exhibiting a slow catalytic rate and poor discrimination between CO2 and O2. O2-fixation leads to photorespiration, which consumes energy and releases fixed C O2. Accordingly, Rubisco catalysis often limits the growth rate of photosynthetic organisms, including crop species. We aim to reimagine CO2 fixation in land plant chloroplasts, however, this requires a nuanced understanding of the biogenesis pathway: we have to fully understand the pathway before we can hack it! Rubiscos from higher plants are comprised of eight catalytic large- (LSu, rbcL gene) and eight auxiliary small- (SSu, rbcS gene) subunits, which form a L8S8 hexadecamer. The nucleus of higher plants encodes an rbcS multigene family that may provide the opportunity for differential SSu expression in response to a range of intrinsic and extrinsic cues. All higher plants have one LSu-encoding gene located in the plastome and a variable number (depending on species) of nuclear-encoded SSu isoforms. We are performing experiments in planta and also by using a SynBio expression system to (i) clarify the structure-functional influence of differentially expressed Arabidopsis SSus, and (ii) investigate the mechanism underlying the incorporation of these different SSu isoforms during Rubisco biogenesis.
Faculty advisors:

Laura Gunn
Molecular and genetic analysis of fruit ripening and related nutrient pathways, using tomato as the model system - Giovannoni Lab, BTI
Project description:

Ripening is a process by which the texture, color, flavor, and nutritional content of fruit is enhanced to make the fruit organ uniquely attractive to fruit consuming, seed dispersing organisms (frugivores). These traits contribute to the healthfulness and desirability of the fruit as a food source. Clearly, understanding the molecular processes behind fruit ripening are important in terms of nutrition, but also for practical applications such as transportation and shelf-life. The Giovannoni lab explores molecular and genetic regulation of fruit ripening, related signaling systems and pathways leading to accumulation of nutritional compounds, using tomato as a model. Researchers in the lab have characterized numerous genes related to ripening control. In addition to identifying important genomic and regulatory components of ripening, the lab investigates regulation of carotenoid synthesis and accumulation in fruit. Lycopene and beta-carotene give tomatoes their color with the latter a precursor of vitamin A. Both are suggested to inhibit degenerative diseases such as cancer and heart disease. Using a genomics approach, the lab is investigating the regulatory mechanisms behind accumulation of this and other nutritionally important compounds in addition to ripening control genes.
Faculty advisor: Jim Giovannoni

Biological nitrification inhibition: Plant breeding to reduce nitrogen emissions - Gore Lab, Cornell
Project Description:
Modern agriculture’s dependence on nitrogen input has led to high yields but at the cost of eutrophication of our waterways through nitrate runoff and increased greenhouse gases through nitrous oxide emissions. A potential avenue to combat the environmental impact of nitrogen while improving yields without additional inputs is a process called biological nitrification inhibition or BNI. BNI has been established to lower nitrogen loss through specific plant root exudates that control soil microbial diversity. Yet efforts to breed BNI in crops such as maize, rice, or sorghum are slow, mainly due to the difficulties in assessing BNI capabilities. This research project combines engineering, biology, and data science to link above-ground plant traits to below-ground processes and enhance the pace at which we can select high BNI lines. The current objectives of this project are to assess the efficacy of hyper and multispectral for the prediction of BNI activity in maize and to understand how BNI activity changes throughout time.
Faculty Advisor: Michael Gore

Molecular analyses of arbuscular mycorrhizal (AM) symbiosis - Harrison Lab, BTI
Project description:
 Phosphorus is a critical macronutrient for proper plant growth. While phosphorus deficiencies can be improved by the application of phosphate fertilizers, it is costly, both to the farmer and to the environment. Furthermore, the crops only take up a small percentage of the applied fertilizer; the remainder is either immobilized in the soil, or carried into ground water and rivers, often resulting in pollution.
Phosphorus is a critical macronutrient for proper plant growth. While phosphorus deficiencies can be improved by the application of phosphate fertilizers, it is costly, both to the farmer and to the environment. Furthermore, the crops only take up a small percentage of the applied fertilizer; the remainder is either immobilized in the soil, or carried into ground water and rivers, often resulting in pollution.
Interns in the Harrison lab investigate two aspects of plant phosphorus nutrition. The first aspect seeks to understand the basis for the symbiotic relationships between vascular flowering plants and arbuscular mycorrhizal (AM) fungi. The fungi colonize root cells, gaining access to carbon supplied by the plant, while at the same time mobilizing mineral nutrients from the soil, including phosphorus, to be used by the plant. For this work, the lab uses the model legume, Medicago truncatula and the fungus Glomus versiforme. The Harrison lab also studies how plants find and take up phosphorus from the soil when they do not have these symbiotic relationships with fungi. This work toward understanding the mechanisms of perception and acquisition of phosphorus by plants may eventually lead to a more effective usage of fertilizers.
Faculty advisor: Maria Harrison

Understanding how insects transmit plant pathogens - Heck Lab, USDA
Project description:
 The Heck lab deciphers molecular mechanisms regulating insect transmission of plant pathogens and uses this knowledge to create practical solutions that mitigate vector-borne diseases in agriculture. We use a combination of computational and wet-lab approaches to study vector-pathogen-plant interactions. Students will receive training at the intersections of computational biology, plant pathology, entomology, microbiology, genetic engineering and synthetic biology. Projects involve research on citrus greening disease or diseases caused by aphid-borne poleroviruses, such as the invasive cotton leafroll dwarf virus
The Heck lab deciphers molecular mechanisms regulating insect transmission of plant pathogens and uses this knowledge to create practical solutions that mitigate vector-borne diseases in agriculture. We use a combination of computational and wet-lab approaches to study vector-pathogen-plant interactions. Students will receive training at the intersections of computational biology, plant pathology, entomology, microbiology, genetic engineering and synthetic biology. Projects involve research on citrus greening disease or diseases caused by aphid-borne poleroviruses, such as the invasive cotton leafroll dwarf virus
Faculty advisor: Michelle Heck
Molecular genetic studies of temperature responses and immune responses in plants - Hua Lab, Cornell
Project description:

Proper responses to environmental signals are essential for plant growth, reproduction, and fitness. Understanding the molecular genetic basis of such responses is not only fundamental to the central biological question of signaling and adaptation, but also better prepares us for global climate changes.
Current research projects in Hua lab include 1) identifying critical regulators of chilling tolerance in maize, 2) investigating the regulation of calcium homeostasis and its impact on plant development and immunity, 3) revealing how a family of calcium binding proteins regulate immunity and temperature stress tolerance, and 4) identifying molecular basis for natural variations in freezing tolerance in plants.
Faculty advisor: Jian Hua

Genetic and biochemical mechanisms of plant defense against insects - Jander Lab, BTI
Project description:
 Plants in nature are subject to attack by wide variety of caterpillars, beetles, aphids, and other insect herbivores. Although there are a million or more species of herbivorous insects, any individual plant species is resistant to the vast majority of these. Insect feeding is inhibited by an array of chemical defenses that exhibits great variability both within and among different plant species. However, although it is known that any plant leaf contains several thousand different metabolites, most of these remain unidentified. In the Jander lab we are investigating natural variation in the herbivore resistance of maize, tomato, and potato to elucidate the molecular basis of plant defense traits. Through a combination of genetic crosses, gene expression assays, metabolite profiling, and insect growth experiments, we are able to identify specific plant genes, biosynthetic pathways, and metabolites that are required to mount an effective anti-herbivore defense.
Plants in nature are subject to attack by wide variety of caterpillars, beetles, aphids, and other insect herbivores. Although there are a million or more species of herbivorous insects, any individual plant species is resistant to the vast majority of these. Insect feeding is inhibited by an array of chemical defenses that exhibits great variability both within and among different plant species. However, although it is known that any plant leaf contains several thousand different metabolites, most of these remain unidentified. In the Jander lab we are investigating natural variation in the herbivore resistance of maize, tomato, and potato to elucidate the molecular basis of plant defense traits. Through a combination of genetic crosses, gene expression assays, metabolite profiling, and insect growth experiments, we are able to identify specific plant genes, biosynthetic pathways, and metabolites that are required to mount an effective anti-herbivore defense.
Faculty advisor: Georg Jander

The chemical ecology of plant communication - Kessler Lab, Cornell
Project description:
 Our Lab studies the chemical and molecular mechanisms, ecological consequences and evolutionary aspects of plant induced responses to herbivory. As a specific focus, the group tries to understand how plant chemistry functions as a vehicle of information transfer between organisms and so affects ecological and evolutionary processes. The group’s research includes a number of basic and applied study systems in North America, Kenya, Peru, Costa Rica, and Colombia.
Our Lab studies the chemical and molecular mechanisms, ecological consequences and evolutionary aspects of plant induced responses to herbivory. As a specific focus, the group tries to understand how plant chemistry functions as a vehicle of information transfer between organisms and so affects ecological and evolutionary processes. The group’s research includes a number of basic and applied study systems in North America, Kenya, Peru, Costa Rica, and Colombia.
Locally we work on the chemical ecology of tall goldenrod, Solidago altissima, with a specific focus on plant communication and how chemical information affects population and community dynamics. More recently, we have started to understand how soil microbial communities affect plant chemistry and so the plants’ interactions with other organisms.
 Moreover, the lab studies ways of applying chemical ecological mechanisms for more sustainable pest control and preservation of ecosystem functions in agriculture.
Moreover, the lab studies ways of applying chemical ecological mechanisms for more sustainable pest control and preservation of ecosystem functions in agriculture.
Research in the lab typically involves field and laboratory bioassays with plants, insects and microbes as well as chemical analytical methods.
Faculty Advisor: André Kessler

PROGRAMMABLE PLANT SYSTEMS REU project: Social, ethical, and public engagement issues - Lewenstein Lab, Cornell
Project description:
The Social, Ethical, and Public Engagement (SEPE) component of CROPPS is an integral part of the overall project, providing a way to include social systems in the overall set of genomic, plant, soil, climate, and technological systems that shape research on digital biology. The goal of the SEPE component is to identify social and ethical issues and facilitate public engagement around those issues. Public engagement goes beyond “outreach,” and includes listening to concerns expressed by multiple stakeholders and public audiences, then feeding those concerns back into the research process. For example, as rural communities identify limited bandwidth as a concern both for the operation of digital biology and for broader social progress, researchers might design networked systems that include social support beyond their more specific needs.
Faculty advisor: Bruce Lewenstein
Seed-free plant genomics and symbioses– Fay-Wei Li Lab, BTI
Project description:
 The Li lab uses genomic tools to investigate the evolution and biology of seed-free plants: ferns, fern allies, and bryophytes (mosses, liverworts, and hornworts). We are particularly interested in their symbioses with N2-fixing cyanobacteria, which provide plants with their own source of nitrogen fertilizer. We aim to understand the evolution and mechanisms of plant-cyanobacteria symbioses, and the results of which will lay the foundation for future engineering of “self-fertilizing” crops with less reliance on synthetic nitrogen fertilizers. To this end, we are surveying the diversity of cyanobacterial symbionts in hornworts, and using RNA-sequencing to uncover putative symbiosis-related genes.
The Li lab uses genomic tools to investigate the evolution and biology of seed-free plants: ferns, fern allies, and bryophytes (mosses, liverworts, and hornworts). We are particularly interested in their symbioses with N2-fixing cyanobacteria, which provide plants with their own source of nitrogen fertilizer. We aim to understand the evolution and mechanisms of plant-cyanobacteria symbioses, and the results of which will lay the foundation for future engineering of “self-fertilizing” crops with less reliance on synthetic nitrogen fertilizers. To this end, we are surveying the diversity of cyanobacterial symbionts in hornworts, and using RNA-sequencing to uncover putative symbiosis-related genes.
Faculty advisor: Fay-Wei Li

Pollinator health - pesticide, pathogen, and nutritional stress on bees - McArt Lab, Cornell
 Project description:
Project description:
Why are pollinator populations declining and what can we do about it? These are core research motivations in the McArt lab. Some current projects include: 1) Understanding pathogen transmission in plant-pollinator networks. We’ve recently found that ~20% of individual flowers have bee pathogens on them (!) and are working to understand how disease spreads in bee communities via bee-flower visitation networks. 2) Investigating how fungicides impact bees during pollination of apple. We’re unraveling a complex system where interactions between fungicides, insecticides, bee microbiota, and pathogens are all at play. 3) Understanding how pollinator populations respond to mass flowering events in agricultural systems and habitat enhancements (e.g., large wildflower plantings underneath solar panels at new solar power facilities).
Approaches in our lab typically involve field and/or lab research with bees, chemical analyses (HPLC, etc.), and molecular techniques (PCR, etc.).
Data-driven approaches to unravel and engineer plant biochemistry - Moghe Lab, Cornell
Project Description:
 The Moghe Lab is fascinated by the diversity in plant metabolites and the genetic and evolutionary mechanisms that create it. Given the rapid pace of new data generation in today’s big data world, we seek to use a combination of bioinformatic and biochemical approaches to unravel plant metabolism, in species such as tomato, sweet potato and Brachypodium distachyon. Students interning in the lab will primarily code in Python and/or R, and will learn advanced genomic, phylogenetic, machine learning and protein structural analysis algorithms. The computational component is supported robustly by experimentation, where we use approaches such as enzyme assays, mutagenesis, and ultrahigh resolution mass spectrometry. Previous summer students in the lab have successfully been co-authors on publications, speaking to the high calibre of students in the REU program and those entering our lab!
The Moghe Lab is fascinated by the diversity in plant metabolites and the genetic and evolutionary mechanisms that create it. Given the rapid pace of new data generation in today’s big data world, we seek to use a combination of bioinformatic and biochemical approaches to unravel plant metabolism, in species such as tomato, sweet potato and Brachypodium distachyon. Students interning in the lab will primarily code in Python and/or R, and will learn advanced genomic, phylogenetic, machine learning and protein structural analysis algorithms. The computational component is supported robustly by experimentation, where we use approaches such as enzyme assays, mutagenesis, and ultrahigh resolution mass spectrometry. Previous summer students in the lab have successfully been co-authors on publications, speaking to the high calibre of students in the REU program and those entering our lab!
Faculty Advisor: Gaurav Moghe

Bioinformatics and genomics - Mueller Lab, BTI
Project Description:
 The Mueller Lab develops software and databases for genomics and genome-based plant breeding with typical intern projects involving analyses and the implementation of tools to facilitate breeding workflows as well as the identification of improved breeding lines.
The Mueller Lab develops software and databases for genomics and genome-based plant breeding with typical intern projects involving analyses and the implementation of tools to facilitate breeding workflows as well as the identification of improved breeding lines.
Faculty Advisor: Lukas Mueller

Defense mechanisms in maize, with a focus on mycotoxigenic fungi - Nelson Lab, Cornell
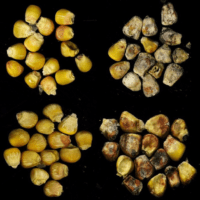
Integrated field, greenhouse and lab studies to reduce the vulnerability of maize to fungi that cause disease
Project description:
Like other crops, maize is attacked by diverse microbes, including micro-fungi that produce toxic compounds. These toxins, including aflatoxin, fumonisin and deoxynivalenol, contaminate staple crops around the world and pose health threats to vulnerable human and animal populations. The Nelson lab is working to understand the genetic architecture of disease resistance in maize and sorghum as well as the mechanisms that enhance or reduce toxin accumulation in crops before and after harvest. We also contribute to plant disease management efforts in international contexts. We work at Cornell, with collaborating labs in North Carolina and Mississippi, and with collaborating teams in India, Tanzania and Kenya.
Faculty advisor: Rebecca Nelson

Dissecting the identity and evolution of meiotic recombination hotspots using genomics and computational biology - Pawlowski Lab, Cornell
Project description:
The goal of the Pawlowski lab is to understand the basis of inheritance in plants by studying the mechanisms governing recombination in meiosis. Meiosis is a specialized type of cell division that leads to the production of gametes. During meiosis, homologous chromosomes, one from the mother and the other one from the father, pair with each other and exchange parts in the process of recombination, which is essential for accurate transmission of genetic material from parents to progeny and for generating genetic variation. Our research combines computational biology as well as genomics methods to identify sites in the genome where recombination takes places and study their evolution. These basic studies provide means to investigating how meiotic processes can be modified to improve plant breeding methods by targeting recombination to desired sites in the genome. .
Faculty advisor: Wojtek Pawlowski

The role of membrane transport in root abiotic stress responses – Pineros Lab, USDA/Cornell
Project description:
Roots are the essential organ for plant nutrition, absorbing water and nutrients. Research in the Pineros lab focuses on the role of two distinct, but complementary aspects of root biology and plant adaptation to environmental stresses: root system architecture and membrane transport. Our goal is to understand the physiological and molecular processes underlying plant abiotic stress responses, as well as mineral nutrition-related processes. Current projects focus on A) structural and functional studies on membrane transporters that underlie Al resistance responses in crops by mediating Al exclusion or internal detoxification, and B) determining the mechanisms underlying the expression and regulation of these membrane transporters. The outcome of this research will provide a new framework for identifying molecular determinants that confer high levels of Al resistance, with the ultimate goal of “engineering” their functional characteristics to enhance the plant’s adaptation responses.
Faculty advisor: Miguel Pineros

Landscape simplification affects plant traits by mediating shifts in the insect community - Poveda Lab, Cornell

Project description:
Our research group studies the ecology of plant-insect interactions in agricultural systems and their interface with natural systems. We focus on two main themes: 1) The effect of diversity at local and landscape scales on ecosystem (dis)services essential for agricultural systems, including pollination, herbivory, biological control, and ultimately yield, and 2) the ecological, physiological, and genetic mechanisms of plant tolerance and resistance traits in agricultural crops. This summer (2022), we are focusing on a project where we are testing how agricultural land-use change alters insect communities and if this results in trait adaptation in local plant populations. Preliminary bioassay data suggest that plants from agriculturally dominant landscapes are more palatable to herbivorous insects and have greater inducibility than plants sourced from landscapes with more natural land cover. Students collaborating on this project will assist with chemical analyses to connect the bioassay results with the plant chemistry to identify the mechanisms driving these patterns.
Faculty advisor: Katja Poveda

Chemical Ecology, Plant-Pollinator Interactions and Multi-Modal Behavior - Raguso Lab, Cornell
Project description:
My students and I study the full spectrum of chemically mediated interactions between flowering plants and their insect pollinators, including typical nectar- or pollen-based systems, obligate mutualism, mimicry and deception. These studies have compelled us to develop broader interests in the evolution of signals and communication, exploring the links between chemical signals and nutrition, physiology and foraging decisions. Our study of chemically-mediated interactions has expanded to include the impact of third parties (microbial symbionts, parasites and predators), and to apply what we have learned in pollination systems to those of host specificity and biological control, including recent work on yeasts, bacteria and the spotted wing drosophila fly.
Our laboratory environment provides students with opportunities to design and perform behavioral bioassays, to learn analytical chemistry, electrophysiology, dietary manipulation and performance assays with several species of insects, plants and microbes. Learn more about our projects, publications and lab history at: https://ragusolab.weebly.com/
Faculty advisor: Robert Raguso

Using microbiomes to edit the plant epigenome and increase plant health - Restrepo Lab, BTI
Project description:
Climate change is making it more difficult to feed the world than it has ever been before. Among other impacts, it has significantly increased the incidence of plant diseases and, as a consequence, of pesticides used to control them. To meet this challenge, one strategy that has earned strength and relevance in the past few years is the use of microbial communities to enhance plant health. In particular, using plant-growth promoting microorganisms to maintain healthy environments and improve plant resistance to pathogens. In many cases, these microorganisms may even be a component of the plant´s natural microbiome! The Restrepo Lab, new to BTI, is led by Dr. Silvia Restrepo who has more than 20 years of research experience in plant pathology and molecular biology. This upcoming summer, we want to explore how microbiomes could be used to improve plant growth and resistance to pathogens. In particular, we will use potatoes as our crop of interest and Phytophthora infestans, the most devastating pathogen of potatoes, as our target. Given that microbiomes have also been known to trigger changes in a plant´s epigenome, we are interested in testing if such changes could specifically improve plant health. Epigenomic marks determine whether a gene is transcribed into protein and the level in which this transcription occurs. Therefore, if microbiomes can be used to modulate gene expression through the epigenome, we could potentially uncover a novel and alternative way to improve plant health.
Faculty advisor: Silvia Restrepo

Nuclear architecture and epigenetic regulation of the genome - Richards Lab, BTI
Project Description:
The Richards lab studies epigenetics – the information content of the genome layered on top of the primary nucleotide sequence. The most intensely studied epigenetic codes are those embedded in the DNA, such as cytosine methylation, or involving post-translational modification of the histone proteins that comprise the nucleosomes. A less-studied aspect of epigenetics is the three-dimensional organization of the genome and the effect of alternate spatial configurations on gene expression. Our studies are focused on this higher level of epigenetic regulation with an emphasis on how nuclear structure impacts, and is shaped by, this regulation. In collaboration with the Lammerding lab at Cornell, we have support from the National Science Foundation to examine how genome size and epigenetic codes, such as cytosine methylation and histone modification, alter nuclear biomechanics. These studies are being carried out using insect cells, as well as plant genetic models
Faculty Advisor: Eric Richards

Cell size and sepal size in Arabidopsis - Roeder Lab, Cornell
Project Description:
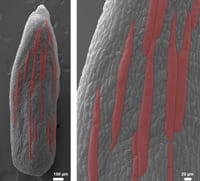 Size is a critical property of plants, yet we know little about how it is controlled. In the Roeder laboratory, we ask how does a cell know how big to be and how does the whole organ (e.g. leaf, sepal or petal) made up of many cells know how big to be. To answer these questions we image living plants on a confocal microscope and measure the growth of cells, examine the cell division pattern, and quantify fluorescent proteins expressed.
Size is a critical property of plants, yet we know little about how it is controlled. In the Roeder laboratory, we ask how does a cell know how big to be and how does the whole organ (e.g. leaf, sepal or petal) made up of many cells know how big to be. To answer these questions we image living plants on a confocal microscope and measure the growth of cells, examine the cell division pattern, and quantify fluorescent proteins expressed.
Faculty Advisor: Adrienne Roeder

Chloroplast biology - Stern Lab, BTI
Project Description:

The Stern laboratory focuses on photosynthetic carbon assimilation and gene expression mechanisms in the chloroplast, where photosynthesis takes place. We use molecular genetic techniques to test hypotheses for increasing plant performance and photosynthetic efficiency by creating and analyzing transgenic plants. Our gene expression work focuses on a key ribonuclease, RNase J, which exerts quality control over the transcript population of the chloroplast, and is itself essential for plant embryo development and viability.
Faculty Advisor: David Stern

Plant-Insect Ecology - Thaler Lab, Cornell
Project Description:
Dr. Thaler’s lab goals are to develop a predictive framework for understanding the complex interactions that occur between plant and insect species. Studies of fundamental ecological processes, in both agricultural and wild systems, can provide insight into controlling insect pests and understanding the natural world. Thaler’s research focuses on ecological interactions between plants, herbivores, and carnivores in agricultural and wild Solanaceous plants. Current research projects focus on understanding the non-consumptive effects of predators on prey; how plants balance interactions between mutualists and antagonists such as pollinators and herbivores, and understanding how plants integrate their defenses against multiple attackers.
Faculty Advisor:
Jennifer Thaler

In Planta (tissue-culture free) Plant Bioengineering - Van Eck Lab *Project only available for the Programmable Plant Systems and the internet of living things REU #2019674
Project Description:
Generation of stable transgenic and gene-edited lines has become a major bottleneck that hinders advancement of plant research because the capacity to generate these lines cannot keep pace with the demand. Bottlenecks can result from several factors such as recalcitrance of species to plant tissue culture methods, low plant regeneration and transformation efficiencies, and inefficient delivery methods of plasmid DNA into plant cells. By far, the need to rely on plant tissue culture methods presents the biggest hurdle for plant bioengineering because the methods are laborious and time consuming. In addition, plant tissue culture requires specialized skills and equipment that limit the number of labs that can be involved in plant bioengineering.
Therefore, the focus of this project is to explore tissue-culture free or in planta methods that allow engineering at the whole plant level. In-planta methods will broaden the number of research groups who can do plant bioengineering because tissue culture-related skills and equipment are not needed. The ability to do whole plant engineering will significantly open the pipeline for recovery of stable transgenic and edited lines not only for the Center for Programmable Plant Systems (CROPPS) project, but also the broader plant research community.
Faculty Advisor:
Joyce Van Eck

Protein degradation in chloroplasts; determinants of the life-time of chloroplast proteins - Van Wijk Lab, Cornell
Project Description:
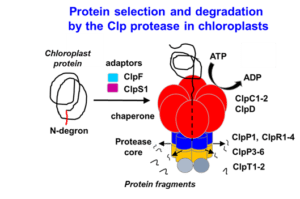
Complexity increases as you follow along the “central dogma” of biology. Model plants, such as Arabidopsis thaliana, have ~27,000 protein-coding genes (DNA) which encode several-fold more transcripts (RNA), which in turn encode hundreds of thousands of proteins. Further, proteins can be chemically modified to yield even greater complexity. Understanding how plants, or any organism, can manage such daunting complexity at the protein level requires the study of protein synthesis, maintenance, modification, and removal; processes that collectively constitute “proteostasis”. The van Wijk lab primarily focuses on the process of (selective) protein removal, called proteolysis. We focus on proteolysis in the plastid; an organelle found in al photosynthetic eukaryotes, including diatoms, algae, crops and trees but also in malaria-causing parasites (Plasmodium). Working with plastids (chloroplasts) is advantageous because of its reduced proteome complexity (~10% of the entire plant proteome) and because the mechanisms of protein degradation are distinct from that of the well-studied proteasomal degradation system in the cytosol of plants. We devote most of our resources towards studying the “natural” substrates and substrate selection mechanisms of the CLP Protease in the model plant species Arabidopsis thaliana, the most abundant protease in plastids. We employ a wide range of techniques, including molecular biology and engineering in Arabidopsis, phenotyping, protein biochemistry, mass spectrometry, recombinant technology in E. coli and bioinformatics. Broadly, we expect that an improved understanding of plastid proteolysis could lead to meaningful technologies with applications in agriculture and medicine.
Faculty Advisor: Klaas Van Wijk

Long distance micronutrient signaling and their role in reproduction and seed nutritional quality in plants– Vatamaniuk Lab, Cornell.
Project Description:
The global demand for high-yield grain crops is increasing due to the current trend of population growth, global climate change, and environmental pollution. In this regard, micronutrients such as iron and copper are required for the growth and development of all organisms including plants and humans. These elements, however, are toxic when are accumulated in cells in access. Thus plants tightly regulate copper and iron uptake from the soil to avoid deficiency while precluding toxicity. This regulation involves the transcriptional control of genes mediating copper and iron uptake from the soil, root-to-shoot partitioning and shoot-to-root signaling of copper and iron status to accommodate the demands of the growing shoot. Many of the mechanisms involved in metal transport, its regulation and signaling as well as micronutrient utilization for ensuring successful developmental programs including fertility are not well understood.
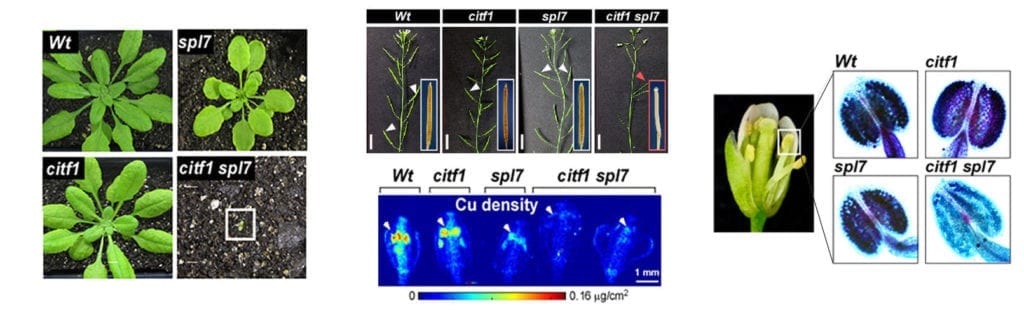
Project 1: Copper transport, it’s regulation, and influence on pollen fertility.
Using RNA-seq analysis, we identified a novel transcription factor, CITF1 (Cu-deficiency Induced Transcription Factor1), that is strongly upregulated in Arabidopsis thaliana flowers subjected to copper deficiency. We demonstrated that CITF1 regulates copper uptake into roots and delivery to flowers and is required for normal plant growth under copper deficiency. We found that CITF1 acts together with a master regulator of copper homeostasis, SPL7, and the function of both is required for copper delivery to anthers and pollen fertility. We now aim to identify the sites of copper action in anthers and pollen, the role played by SPL7 and CITF1 in pollen development, SPL7, and CITF1 transcriptional regulatory networks and transport processes governing copper homeostasis in A. thaliana. We are also analyzing SPL7 and CITF1 pathways in a globally important crop, wheat, and its proxy, a model grass species, Brachypodium distachyon.
Project 2: Shoot-to-root signaling of iron deficiency
Despite significant progress in the understanding of how plants acquire iron from the soil and how iron is mobilized within the plant, not much is known about how shoots communicate their iron status to the root. We are using A. thaliana iron deficiency signaling mutants to address the question of the nature of systemic iron deficiency signal(s), its interactions with sensors in different tissues and cell types as well as the signal propagation to root epidermal cells to trigger transcriptional iron deficiency responses.
Faculty Advisor: Olena Vatamaniuk

Internships are funded by the National Science Foundation, Research Experiences for Undergraduates Award #1358843, individual faculty grants, and the generosity of donors including the Emerson Foundation , Ithaca Garden Club, John Ben Snow, the Legacy Foundation of Tompkins County, Rheonix, Triad Foundation Inc, Yunis Realty , and many individual donors.