Fay-Wei Li

We are broadly interested in the evolutionary processes at the gene, genome, and microbiome levels that shaped the plant diversity. We mostly focus on “seed-free” plants (ferns, lycophytes, and bryophytes), and anything that has a weird biology. Check out my lab website for details.
Evolutionary genomics of seed-free plants
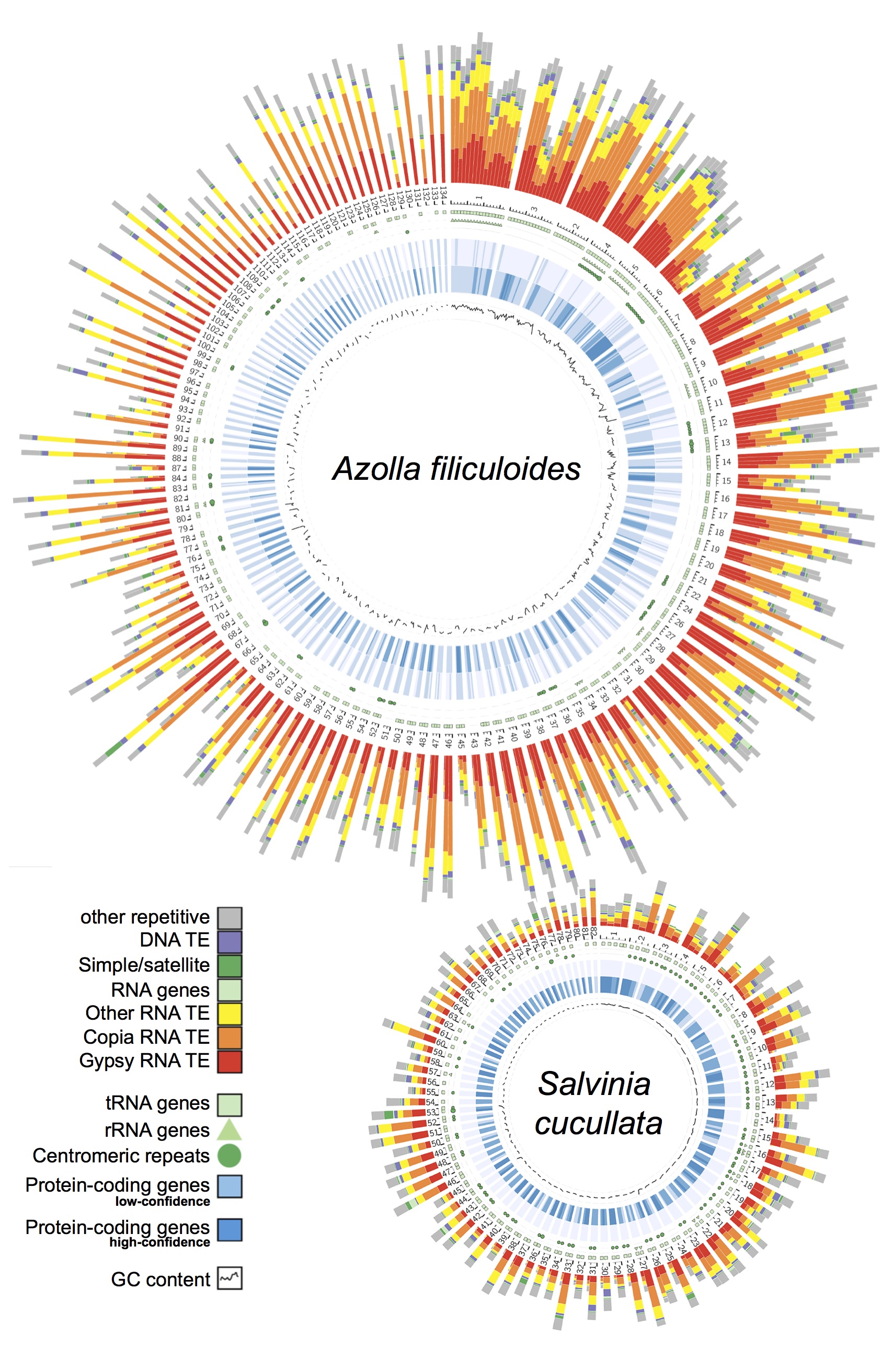
Genomic information is critical to elucidating the evolutionary history and the genetic basis of diverse traits. However, despite over 300 plant genomes having been published, the majority of them were from seed plants and very little is known about seed-free plants. Our group is leading the effort to not only close this large “genome gap”, but also leverage omics data to understand the unique biology of seed-free plants. To date, we have completed the first genomes for ferns, hornworts, and Isoetales, and have more in the pipeline.
Ferns—In 2018, our group led an international collaboration to complete the first genomes of ferns (Li et al., 2018). These genomes, from Azolla and Salvinia, not only provided an exciting glimpse into fern genome structure and evolution, but also clarified the origins of gene families and pathways that coincide with the emergence of seed plants. We have continued to play a lead role in advancing fern genomics. In 2022 we published the genome of the tree fern Alsophila spinulosa (Huang et al., 2022). This genome is notable because Alsophila is homosporous (in contrast to the heterosporous Azolla and Salvinia) and the size, at 6 Gb, is more typical of ferns. We discovered a remarkably preserved synteny, despite resulting from an ancient whole-genome duplication over 100 million years ago. Such a slow rate of genome fractionation is unprecedented and might explain the large genome sizes found in ferns. In parallel, we have also been working closely with Open Green Genomics (a JGI-funded project) and aim to have four more fern genomes completed by the end of 2022.
Lycophytes—There are three extant and deeply diverged lineages in lycophytes: Selaginellales, Isoetales, and Lycopodiales. For years, genomic data had only been available for Selaginella in Selaginellales. Recently, we made important progress in lycophyte genomics with the completion of Isoetes taiwanensis (Isoetales) genome (Wickell et al., 2021). Our interest in I. taiwanensis is primarily in its Crassulacean acid metabolism (CAM) photosynthesis, which is unusual given the aquatic lifestyle. From our time-course RNA-sequencing data, we found that I. taiwanensis may have recruited the lesser-known “bacterial-type” phosphoenolpyruvate (PEP) carboxylase, along with the “plant-type” exclusively used in other CAM and C4 plants for carboxylation of PEP. Our findings suggest the existence of more evolutionary paths to CAM than previously recognized. For the remaining order Lycopodiales, we have finalized the genomes of Diphasiastrum complanatum and Huperzia asiatica, which, together with those of Selaginella and Isoetes, will give us a much more holistic view of the lycophyte genomic landscape.
Hornworts—Hornworts are one of the three lineages of bryophytes, whose phylogenetic position among land plants has been highly debated. In 2020 we published the first hornwort genomes from two Anthoceros species (Li et al., 2020). Our genomic analyses provided support for the monophyly of bryophytes and help address several long-standing questions on early land plant evolution. Since then, we have massively expanded the sampling, with 10 new high-quality genomes from every hornwort family—hornworts are now the best sampled plant phylum to date. This genomic framework is paving the way to establish hornworts as a new model lineage.
Hornworts as a new model system
Hornworts are distinct in many aspects. Phylogenetically, hornworts are one of the major plant lineages that diverged from other extant land plants over 480 million years ago. As such, hornworts are of paramount importance to retrace plant evolutionary history. Physiologically, hornworts have endophytic symbiosis with mycorrhizal fungi and N2-fixing cyanobacteria, pyrenoids for concentrating CO2, a chimeric photoreceptor, and U/V sex chromosomes. Developmentally, hornwort sporophytes are unique in having a basal semi-indeterminate meristem and stomata that are perpetually open. Research on hornworts can therefore bring novel insights into a wide range of biological disciplines, such as plant evolution, evo-devo, plant-microbe interactions, and carbon fixation. In order to facilitate hornwort research, we have been building several key resources and foundational tool:
Collections of cultures and genomes—We have established a diverse hornwort culture collection that includes 24 isolates from 10 genera representing all 5 families, some of which have already been fully sequenced. The cultures are axenic and derived from a single spore or thallus. Importantly, this culture collection contains 6 pyrenoid-present and 8 pyrenoid-absent species, which together cover 3 independent transitions over the hornwort phylogeny. In addition, Peter Schafran (NSF Plant Genome Postdoc) is now expanding this collection with separate male/female isolates to study hornwort sex chromosomes.
Genetic tools—With funding from the NSF EDGE program (to me and Joyce Van Eck at BTI), we have been developing gene delivery and gene editing methods. In collaboration with Eftychis Frangedakis (Cambridge), Keiko Sakakibara (Tokyo), and Peter Szövényi (Zurich), we published the first Agrobacterium-mediated transformation protocol for hornworts (Frangedakis et al. 2021). In parallel, we have been testing a biolistic delivery method, which we believe is more efficient and better suited for CRISPR-Cas9 gene-editing. In one bombardment, we can now reliably obtain ~900 transiently transformed events from 0.3 g of tissue. CRISPR mutagenesis experiments are also underway. Finally, a new growth medium for hornworts was developed that yields >4X tissue mass compared to the original formula (Gunadi et al., 2022).
Supported by NSF IOS EDGE | Developing transformation capacity for Anthoceros agrestis to facilitate gene function studies in hornworts, a remarkable phylum of plants. PI: Fay-Wei Li, Co-PI: Joyce Van Eck (BTI)
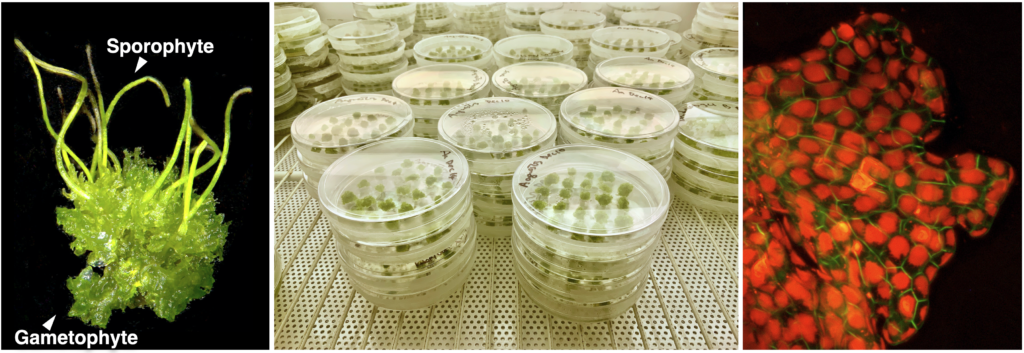
Hornwort-cyanobacteria symbiosis
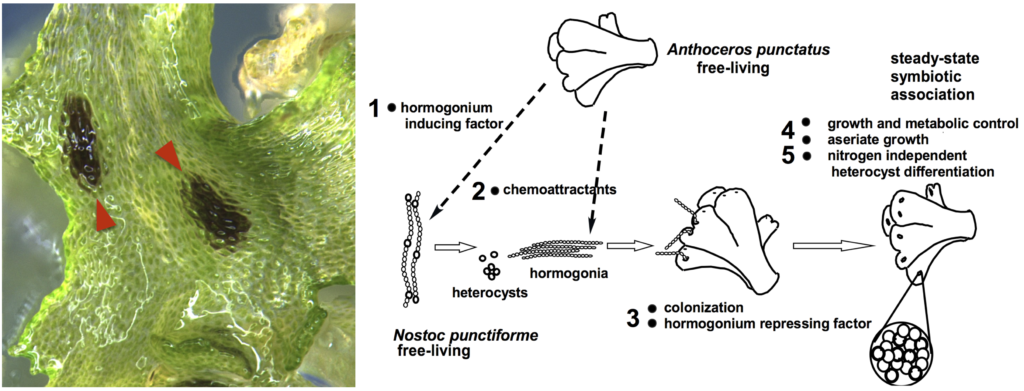
Plant symbiosis with nitrogen-fixing cyanobacteria is a unique form of mutualistic association that has independently evolved in diverse lineages including a few species of bryophytes, ferns, cycads, and one small genus of flowering plants. Compared to other nitrogen-fixing microbes, cyanobacteria are generally less dependent on the plant host, and therefore could be an ideal partner for engineering symbiotic nitrogen fixation into crop plants. However, our current understanding of plant-cyanobacteria symbioses is rudimentary. The phylogenetic diversity of cyanobionts has been largely unexplored, and very few studies have investigated variation in the symbiotic interaction. In addition, most genetic research has solely focused on the model cyanobiont Nostoc punctiforme, and the plant genes involved in symbiosis remain unknown.
Hornworts are an ideal system to study such symbiotic interactions, primarily due to the ease of manipulation in the laboratory and the groundwork laid by us and our collaborators. With a five-year grant from NSF’s Dimensions of Biodiversity program, we are starting to gain a much better understanding of hornwort-cyanobacteria symbiosis.
Ecology and Evolution—To characterize the diversity of symbiotic cyanobacteria in hornworts, we have developed a new PacBio-based amplicon-seq method. We carried out repeated samplings of hornworts and soils in Central New York. Three sympatric hornwort species were included, allowing us to directly compare partner selectivity. We found that hornwort cyanobionts are phylogenetically diverse, have a tight connection to the soil, but do not appear to be heavily structured by time nor host species. This work represents the most comprehensive ecological study to date on plant-cyanobacteria interaction (Nelson et al., 2021). Going forward, we plan to co-inoculate pure cultures of both partners in highly controlled microcosm environments to tease apart factors that determine the symbiotic outcome.
Our research into the diversity of symbiotic cyanobacteria has also led to a few fortuitous discoveries. While isolating cyanobacteria from field-collected hornworts, we have discovered several genera new to science. The most notable isolate, which we named Anthocerotibacter panamensis, is a member of Gloeobacteria, an enigmatic lineage sister to the crown cyanobacteria. We found A. panamensis lacks thylakoid and diverged from the closest culturable species over 1.4 billion years ago. Importantly, A. panamensis exhibits a suite of unique morphological, physiological, and genomic features, thus offering a unique window to reconstruct the early evolution of Cyanobacteria. This research was selected as the cover story in Current Biology (Rahmatpour et al., 2021).
Physiology—To understand the genetic basis of symbiosis, we have identified a suite of promising candidate genes from hornworts through RNA-seq experiments (Li et al., 2020; Chatterjee et al., 2022). These include a SWEET transporter and a few receptor kinases and transcription factors. We are in the process of characterizing these genes in detail with our new transformation methods. We aim to use promoter-GUS experiments to validate whether the genes are predominantly expressed in cells surrounding the cyanobacterial colonies. We then intend to generate CRISPR knockout alleles of validated targets to study the functional consequences of these genes and their products.
Supported by NSF DEB Dimensions of Biodiversity | Integrating phylogenetics, ecophysiology, and transcriptomics to understand the diversity of hornwort-cyanobacterium symbiosis. PI: Fay-Wei Li, John Meeks (University of California Davis), Jed Sparks (Cornell EEB)
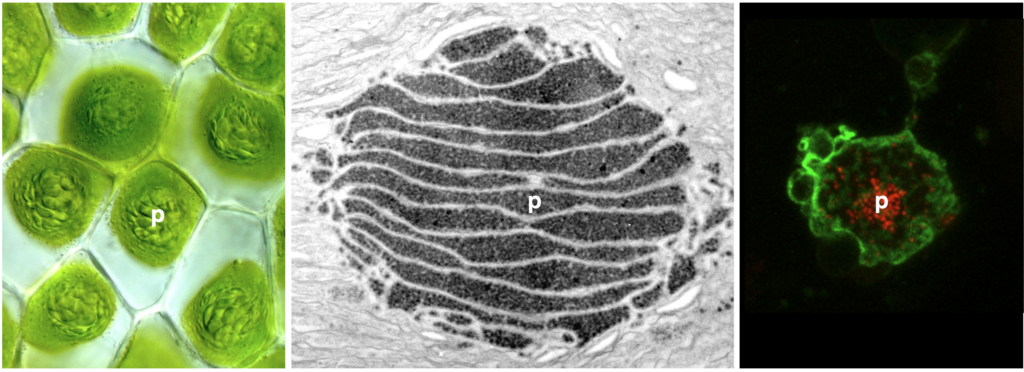
Carbon-concentrating mechanism in hornworts
To enable more efficient photosynthesis, certain organisms have evolved biophysical carbon concentrating mechanisms (CCM) that partition Rubisco (nature’s carbon fixing enzyme) and CO2 into a dedicated subcellular compartment. Pyrenoids are an example of such compartments—organelles comprised of an interconnected matrix of aggregated Rubisco that are liquid-liquid phase separated from the stroma. The green alga Chlamydomonas reinhardtii has been the traditional model to study pyrenoid-based CCMs with the hope of installing a similar mechanism into crops to enhance yield. On the other hand, hornworts are the only land plants that have a pyrenoid-based CCM. Owing to their much closer relationship to crop plants, lessons learned from hornworts might have higher translational potential than those from Chlamydomonas. Another advantage of the hornwort system is that CCMs have been repeatedly gained and lost over the past 50 million years in the evolution of this plant group, offering a unique phylogenetic replication for comparative studies.
To date, no hornwort CCM component has been characterized and nothing is known about what mediates Rubisco aggregation in hornwort pyrenoids. Together with Laura Gunn’s group at Cornell University, we recently secured a collaborative NSF grant to study the hornwort CCM. The plan is to use a three-pronged approach—combining comparative genomics, RNA-seq, and proteomics—to identify putative CCM components, leveraging repeated pyrenoid-present/absent transitions to narrow in on candidates. Ultimately, we aim to use a constructionist approach to characterize interactions between Rubisco and recombinantly-produced putative linker proteins in vitro, and assess the potential for Rubisco phase separation. The outcomes of this research should provide clues as to how a biophysical CCM can be assembled in land plants.
Supported by NSF MCB Cellular Dynamics and Function | From phylogeny to biomolecules: a cross-scale approach to understand the making of a unique carbon-concentrating mechanism in hornworts. PI: Fay-Wei Li, Laura Gunn (Cornell Plant Biology)
-
BTI promotes faculty member Fay-Wei Li
The Boyce Thompson Institute (BTI) is delighted to announce that faculty member Fay-Wei Li has been promoted to Associate Professor on January 13. Li was evaluated on his achievements to […] Read more » -
New species of alga named for poet Amanda Gorman
“And so we lift our gaze, not to what stands between us, but what stands before us.” – Amanda Gorman, from “The Hill We Climb.” In 2020, a group […] Read more »
Seed-free plant genomics and symbioses
 The Li lab uses genomic tools to investigate the evolution and biology of seed-free plants: ferns, fern allies, and bryophytes (mosses, liverworts, and hornworts). We are particularly interested in their symbioses with N2-fixing cyanobacteria, which provide plants with their own source of nitrogen fertilizer. We aim to understand the evolution and mechanisms of plant-cyanobacteria symbioses, and the results of which will lay the foundation for future engineering of “self-fertilizing” crops with less reliance on synthetic nitrogen fertilizers. To this end, we are surveying the diversity of cyanobacterial symbionts in hornworts, and using RNA-sequencing to uncover putative symbiosis-related genes.
The Li lab uses genomic tools to investigate the evolution and biology of seed-free plants: ferns, fern allies, and bryophytes (mosses, liverworts, and hornworts). We are particularly interested in their symbioses with N2-fixing cyanobacteria, which provide plants with their own source of nitrogen fertilizer. We aim to understand the evolution and mechanisms of plant-cyanobacteria symbioses, and the results of which will lay the foundation for future engineering of “self-fertilizing” crops with less reliance on synthetic nitrogen fertilizers. To this end, we are surveying the diversity of cyanobacterial symbionts in hornworts, and using RNA-sequencing to uncover putative symbiosis-related genes.





















