The following is a guest post from Savanah Dale, a Plant Breeding and Genetics Ph.D. student at Cornell University, as part of the Approaches for Crop Improvement Blog Series. She can be reached at smd346@cornell.edu.
Welcome back to the third installment of the Approaches for Crop Improvement blog series! Previously, we discussed the merits of traditional plant breeding (post #1) and mutagenesis (post #2), which people use to produce improved plant varieties for applications such as food production. Although traditional breeding has enabled us to create the vast majority of the crops we know and love, it has many constraints, such as the time it takes for new genetic variation to arise, and the scarcity of that unique variation. Here mutagenesis has benefited the world of plant breeding by providing a means to increase the number of mutations that contribute to new variation over a relatively short amount of time. Yet there is still room for more improvement; even though we are able to increase the diversity of plant traits through mutagenesis, we cannot produce precise results using this method due to the complex nature of the genetic code (DNA). We cannot know exactly which traits a specific mutation is affecting and how it is affecting them unless we know exactly which DNA sequences control those traits. We want to know, if we induce mutation in the genetic code in a specific region, will it cause the fruit color to change to red or pink? Or will it produce some other physical change entirely? Genomic sequencing allows us to predict these changes, or at least understand them. With this technology, plant breeders can detect which DNA nucleotides (each subunit of DNA represented by the letters A, T, C, and G) make up a gene, and can therefore observe which physical characteristics correlate with that particular gene. Most spectacularly, genomic sequencing, which is sequencing of the entire genetic sequence, or genome, of a plant, can be read all at once in order to compose a complete record of all of the genes in that plant, as well as the different alleles, or gene versions. This ability has opened the door to an entire generation of advancement in plant breeding success.
History and development of genomic sequencing
The first sequencing in history was not of DNA, but of proteins, conducted by a biochemist, Fred Sanger, in the early 1950s. Proteins are made up of sequences of amino acids, which are compounds sometimes referred to as the building blocks of life. This first generation, or Sanger sequencing, method consists of cutting up proteins into random small fragments that have overlapping nucleotide sequences at their ends, and then using those overlapping ends to put the fragments back into the correct order of the original full-length sequence. It is much easier to read the amino acid sequences of these smaller fragments than the entire protein at a single time, but then you have to figure out how to put them in order again. Imagine having several strips of paper with the same sentence written on them. If you used scissors and randomly cut those sentences into fragments, you would end up with a bunch of random words that don’t make sense. However, you could line up fragments that share common words to figure out what the original sentence was (Figure 1). This is the basic function of sequencing. 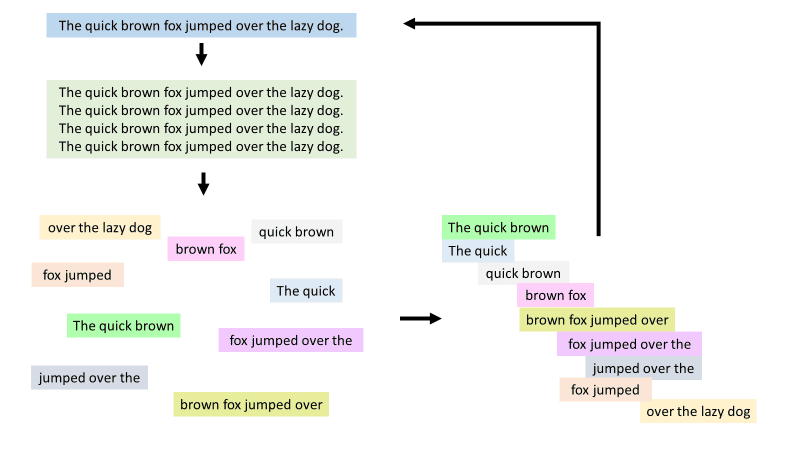 Figure 1. If you chop up a sentence into random pieces that are then jumbled up, you can use overlapping words to figure out the original sentence. Source: Savanah Dale
Figure 1. If you chop up a sentence into random pieces that are then jumbled up, you can use overlapping words to figure out the original sentence. Source: Savanah Dale
In 1976 the first methods for DNA sequencing were invented, which was a natural progression from sequencing proteins because each protein is generated from its own unique genetic code or DNA sequence. Behind every great protein, there is a great DNA sequence. Just as proteins are made up of sequences of amino acids, DNA is made up of sequences of nucleotides (A, T, C and G). In order to sequence nucleotides, Sanger and Coulson developed the chain terminator method, and Maxam and Gilbert developed the chemical cleavage method. Both of these methods worked similarly to the protein sequencing methods developed by Sanger. They had different methods of creating short fragments of DNA, but both then used overlapping end nucleotide sequences to order the fragments correctly and determine the entire DNA sequence (Figure 2), similarly to how the overlapping words were used to determine the whole sentence in Figure 1. In 1979, Staden developed the Shotgun sequencing method, which functioned similarly to the previous methods but used a different approach to generate the short DNA fragments from the original sequence. 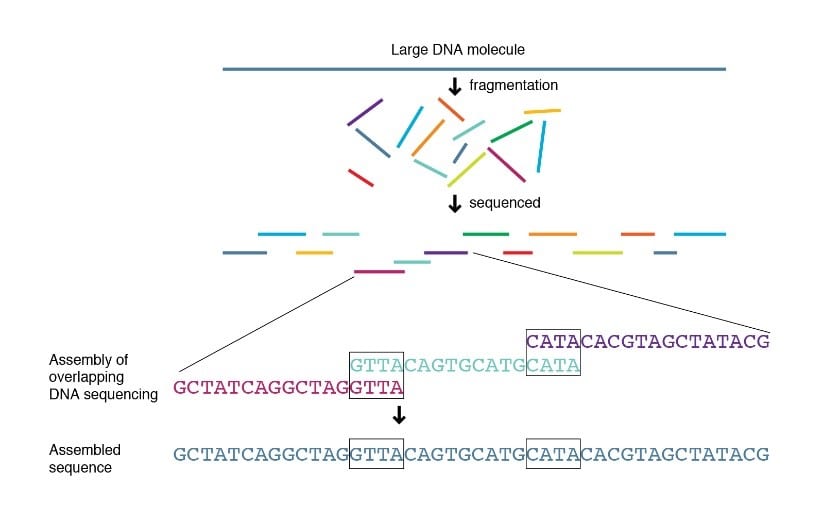 Figure 2. Shotgun sequencing uses overlapping nucleotide sequences on the ends of short DNA fragments to determine the relative order of those fragments in a long DNA sequence. Source: https://www.genome.gov/genetics-glossary/Shotgun-Sequencing
Figure 2. Shotgun sequencing uses overlapping nucleotide sequences on the ends of short DNA fragments to determine the relative order of those fragments in a long DNA sequence. Source: https://www.genome.gov/genetics-glossary/Shotgun-Sequencing 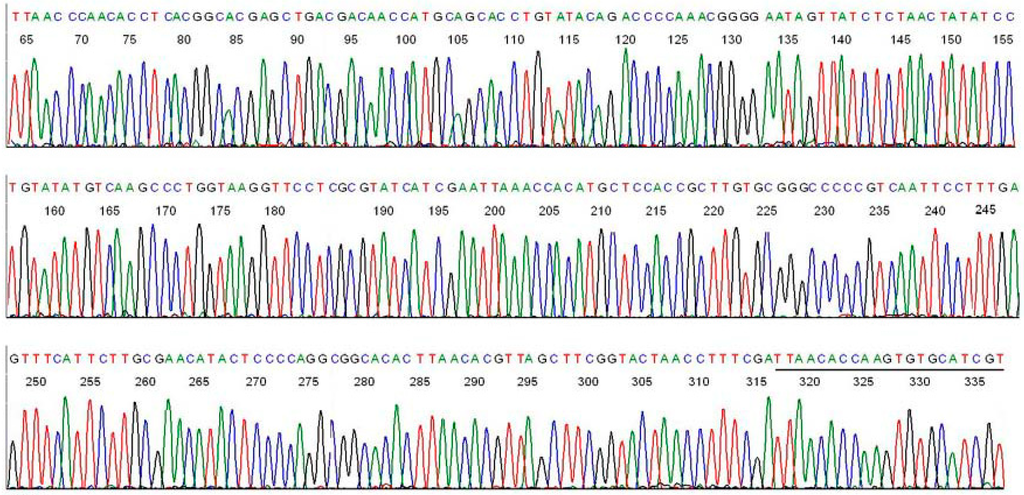 Figure 3. Visual representation of Sanger sequencing data, showing different colored peaks representing different DNA nucleotides (A – green, C – blue, T – red, and G – black). The different nucleotides emit unique chemical signals, allowing them to be “read” by special equipment. Source: https://www.mdpi.com/1422-0067/15/7/11364
Figure 3. Visual representation of Sanger sequencing data, showing different colored peaks representing different DNA nucleotides (A – green, C – blue, T – red, and G – black). The different nucleotides emit unique chemical signals, allowing them to be “read” by special equipment. Source: https://www.mdpi.com/1422-0067/15/7/11364
Over time, machines were developed that increased the speed of Sanger sequencing and the amount of DNA that could be sequenced in a day (1,000 nucleotides in one day in 1987). In 2001, sequencing centers could generate 10 million nucleotides each day. This ability to produce larger amounts of sequence data led to the need for improvement in the world of big data, especially storage and processing, and the amounts of data produced are still increasing today thanks to advances sequencing!
How genomic sequencing is used for plant breeding
Today, genomic sequencing has a multitude of applications in the world of plant breeding. Firstly, this technology can be used to read the entire genetic code of plant species. In order for us to more fully understand the impact that genomic sequencing has had on plant breeding, first imagine that you have to edit an essay your friend has written. Now, imagine that the essay is in a language that you do not speak. This would prove to be a very challenging, if not impossible task, as you neither understand the vocabulary nor the grammar of that language. This is how difficult it was for plant breeders to make use of the natural, inherent variation in species and variation caused by methods like mutagenesis before the advent of genomic sequencing. But now, imagine that you have suddenly become C3PO from Star Wars, and you now speak this language; you can now easily edit your friend’s essay. In the same way, genomic sequencing of plant DNA has opened the doors for traditional breeding and other crop improvement approaches such as gene editing by CRISPR. With this incredible ability, we can gain a much better understanding of the genetics that control a desirable trait in the crops that we want to improve, and a plethora of modern science has evolved from this. Now that we can sequence the entire genome for an individual of a species, we can link the differences between individuals’ physical traits to variation between their genotypes (an individual’s unique genetic sequence). In other words, this technology has allowed us to identify the genes that control specific physical traits, and has allowed us to understand how changing a gene can change the version of the trait that we see. This ability to precisely target a specific gene is known as gene editing. We can also compare the genetic sequence of several individuals to determine how closely related they are, even in enough detail to determine the evolutionary history of a species and where it became different from its ancestors on the tree of life. Additionally, if the entire genomic sequence of an organism is known, then statistical approaches such as Genome Wide Association Mapping can be used to link observed phenotypes to specific genotypes, thus allowing us to develop methods of manipulating many important traits like yield and pest resistance, traits that otherwise would not be possible to easily improve through traditional breeding.
Why there is still room for improvement
With the vast ocean of knowledge provided by genomic sequencing, so many doors in plant breeding are opened. Yet, just the knowledge of what exactly a specific mutation will do in the genome does not entirely solve the challenge of achieving precision in mutagenesis (post #2). This is where gene editing comes into play. This method allows us to create specific desired mutations on demand, and therefore allows a more precise method of plant breeding than has been achievable in the past. Please stay tuned for our next installment to learn how plant breeders can use gene editing for precision breeding!

Savanah Dale smd346@cornell.edu
I am a Plant Breeding and Genetics Ph.D. student at Cornell University, where I am jointly advised by both Dr. Joyce Van Eck and Dr. Michael Gore. I have a B.S. in Plant and Environmental Sciences from Clemson University, where I worked in the Kresovich Lab on sorghum. My interests center around the idea that food systems must provide not only enough calories, but also enough nutrition to their consumers. Therefore, my graduate work is in examining the genetics that control plant specialized metabolites and other phenotypes that are beneficial for human health, as well as how nutrition of vegetable crops can be improved
