2019 Interns Projects and Posters
Interns and Projects (2019):
Connor Lane

Project description:
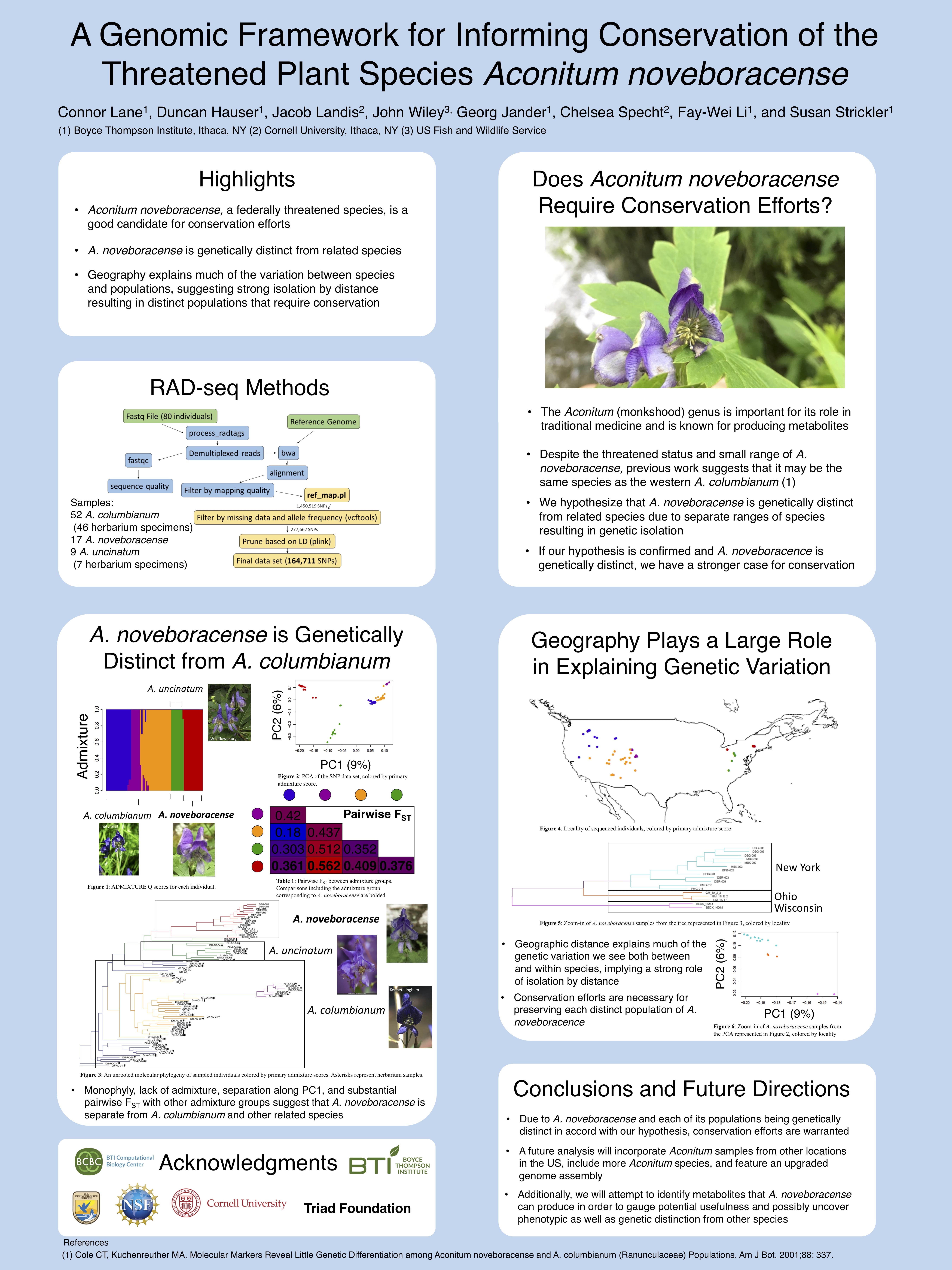
- Aconitum noveboracense, a federally threatened species, is a good candidate for conservation efforts
- A. noveboracense is genetically distinct from related species
- Geography explains much of the variation between species and populations, suggesting strong isolation by distance resulting in distinct populations that require conservation
- Due to A. noveboracense and each of its populations being genetically distinct in accord with our hypothesis, conservation efforts are warranted
- A future analysis will incorporate Aconitum samples from other locations in the US, include more Aconitum species, and feature an upgraded genome assembly
-
Additionally, we will attempt to identify metabolites that A. noveboracensecan produce in order to gauge potential usefulness and possibly uncover phenotypic as well as genetic distinction from other species
Poster:
Tony Kinchen
Project description:
The purpose of our research was to locate tomato genes involved in response to speck, a bacterium known as Pseudomonas syringae,that manifests itself in the form of black spots on the leaves and in drastic cases can cause the leaves to shrivel, wilt, and die. This bacterium is not only limited to tomatoes and has been known to spread to other crops. Our goal for this project was to identify the gene(s) of tomato that are involved in its response to the bacterial speck. Our hope is that this tomato could be improved through breeding a natural resistance to the bacterial speck and aid agriculturalists whose economic stability depends on tomatoes. It is important to note that this research can also be applied to other studies that involve similar circumstances and is not simply limited to tomatoes. Pseudomonas syringae possesses the ability to infect a large variety of plant and crop species aside from tomatoes,meaning the research we acquire in this study could potentially be used to help other crops create plants with a stronger resistance to the bacterium. We hypothesize that by using genetic markers based on primers and PCR we can identify the gene(s) responsible in plants producing a novel (different) response and plants that maintained the wild type response to Pseudomonas. The wild type response typically demonstrates signs of infection through black specks. Novel responses to speck include galls, a swelling of the tissues of the plants, or reduced or increased response.
Poster:
Rohit Lal
Project description:
Bacterial speck is a common disease, caused by the bacterium Pseudomonas syringae pv. tomato, that occurs in tomatoes which is an important agricultural crop. The disease significantly reduces the yield of the crop, therefore not only posing risks on the destruction of crops, but also destroying crops that possibly represent the livelihood of farmers across the world. The disease takes the form of black and brown dots on leaves that spread to the fruit translating to large areas of tissue damage, as well as the wilting of leaves on the fruit. The bacteria easily spreads from plant to plant often through splashing water in the form of rain or watering by sprinkling making it an even bigger problem. All in all, it’s clear that bacterial speck is a major threat to tomatoes in wetter climates, and understanding the growth and conditions that allow for the best growth of the bacteria may help in the fight against disease to improve the production of tomatoes worldwide. Additionally, a standardized and automated method of quantitating disease progression would be invaluable in plant phenotyping.
In this project we used a micro-computer Raspberry Pi to control multiple interfaces through which we were able to create peripherals that combine to create a device that is able to monitor the growth of bacterial speck on plant leaves over time, as well as the conditions that the bacterial speck is growing in. Specifically, we used a camera module that takes pictures over certain intervals (like a time lapse) so we can observe speck growth over time. Also, we have many micro sensors that came in a Raspberry Pi SmartGardenSystem Kit that will allow for the measurement of environmental variables such as temperature, humidity, sunlight, and soil moisture. With this data we will make charts using the R language that will tell us relationships between growth in bacterial speck and its environment. Finally, with the pictures that we collect we will quantitate the disease progression over time using software called ImageJ. The way I will use this software is measuring the amount of total pixels that are seen in the image of the leaves and then I will see how many are a different color (dark green) and then subtracting that from the total. Then I will divide the new total by the reference to get a percentage representing disease progression. The reference image will be the most infected tomato leaves.Then with simple division one can get the progression of the disease in their given experiment. This will become an extremely useful tool for phenotyping tomatoes and can be applied to more applications such as apps to alert farmers of certain stages in disease progression.
Poster: