News
RNase III Discovered to Function in Plant Chloroplasts
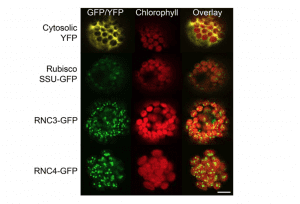
Confocal laser scanning microscopy of Arabidopsis protoplasts transiently expressing the first 100 amino acids of RNC3 or RNC4 fused to GFP. Chlorophyll autofluorescence of chloroplasts is shown in red.
Research Associate Amber Hotto and colleagues, working in the laboratory of BTI President David Stern, have characterized the first RNase III enzymes that function within chloroplasts. They have shown that two such enzymes, classified as Mini-III proteins, can process chloroplast RNA in the model plant Arabidopsis. These processes regulate gene expression within the cells. Their paper, “Arabidopsis Chloroplast Mini-Ribonuclease III Participates in rRNA Maturation and Intron Recycling,” appeared online in February 2015 in the journal The Plant Cell.
RNase III is a unique class of RNA-cleaving enzymes that relies on paired, or double-stranded RNA as a target. This particular property suggests that it should be present in most or all organisms and indeed, advances in genomics have demonstrated that the enzyme is part of a so-called superfamily. Several forms of RNase III are known in bacteria, including Mini-III, and because chloroplasts descended from cyanobacteria that took up residence within another cell more than one billion years ago, bacteria and chloroplasts share many molecular components.
In an earlier publication, Hotto and her colleagues found that chloroplasts contained more than 100 non-coding RNAs, which are not translated into proteins, like messenger RNAs. In other organisms, these non-coding RNAs can turn protein production on or off by stabilizing or destabilizing the mRNAs that carry the information from the genome to the ribosome, among other functions. The researchers thought that if they could find evidence in the chloroplast of an RNase III enzyme—which can regulate non-coding RNAs in bacteria—then the full non-coding RNA circuit might exist in chloroplasts as well.
To do this, the researchers used mutant plants that lacked working genes for the RNase III enzymes in the nucleus. The genome of the model plant, Arabidopsis thaliana, has two genes that code for Mini-III proteins, called RNC3 and RNC4. If the plant was missing just one of these genes, it still grew normally, but if it lacked both, then the plants had insufficient chlorophyll levels and appeared pale green. They were also smaller than wild-type plants.
The researchers isolated chloroplasts from the wild-type and the double mutant plants and extracted the RNA. They then performed RNA-Seq, a high-throughput method that sequences all of the RNAs within a sample, called a transcriptome. They then looked for differences between the two samples that would give evidence of Mini-III protein function, including splicing together specific parts of mRNAs, and RNA processing such as trimming and cutting. “RNA-Seq is quite efficient because in one experiment you have a snapshot of the entire cellular transcriptome,” said Hotto. “You can identify both known and unknown Mini-III targets at once .”
By comparing transcriptomes, they found that the Mini-III proteins are necessary for processing some of the structural RNA components of the ribosomes, certain non-coding RNAs, and also for fully degrading introns—sequences that interrupt mRNAs, which have to be removed and recycled for gene expression.
Knowledge of Mini-III proteins in the chloroplast may aid researchers who wish to control gene expression within the organelle. “The more we know about how chloroplast RNA is metabolized, the more we can think about how genes are expressed, or how we could affect their expression through transgenics, or introduction of foreign genes into the chloroplast,” said Hotto. In a sense, the chloroplast can be a “modifiable bioreactor,” used to produce foreign proteins or lipids of interest in a contained space within a plant cell that is essentially powered by light energy.
Some researchers have demonstrated approaches to express genes that produce vaccine particles within chloroplasts, which could lead to edible vaccines in fruits or vegetables. A project in the Stern laboratory focuses on expressing oil production genes in algae chloroplasts to increase production and potentially yield an alternative source of fuel. While these applications are still far down the road, increasing our understanding of expression and regulation of chloroplast genes can help guide researchers in their initial design phase.


