News
Back to our roots: Insights from genomes of a plant-associated fungus and its bacterial endosymbionts
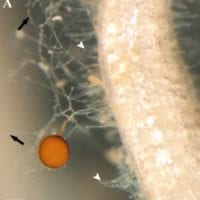
D. epigaea spores (orange sphere) in soil attach to and form associations with plant roots. Tiny root hairs (white arrows) are almost indistinguishable from the network of D. epigaea (black arrows) emerging from the root. A 50 micrometer (0.05 millimeter) bar in the lower left corner is provided for scale.
Image credit: Sun et al., 2018
In an article published this month in the journal New Phytologist, researchers at the Boyce Thompson Institute and the National Center for Genome Resources describe the genome sequences (DNA sequences), of the fungus Diversispora epigaea (formerly known as Glomus versiforme) and its endosymbionts – beneficial bacteria that live inside its cells. D. epigaea is a species of mycorrhizal fungi, which form symbiotic relationships with nearly all land plants. Mycorrhizal fungi are critical for plant health, particularly in nutrient-poor soils. These new genomes help researchers understand the evolution of mycorrhizal fungi and their bacterial endosymbionts.
D. epigaea is at the center of a series of symbiotic relationships. As part of a subset of mycorrhizal fungi called arbuscular mycorrhizal (AM) fungi, D. epigaea colonizes the interior of plant root cells, forming structures called arbuscules. The rest of the fungal body branches out into the soil, providing increased surface area for uptake of nutrients, which it exchanges for energy-rich carbon compounds from its plant host. In addition to its relationship with plants, D. epigaea is also host itself to bacterial endosymbionts, which live inside the fungus. These complex, nesting relationships are not well understood, despite the ubiquity and importance of mycorrhizal fungi.
Organisms that depend on other species for survival typically have smaller genomes than their free-living relatives, as they have ‘outsourced’ many processes to their host. However, this does not appear to be the case for D. epigaea. “The genomes of AM fungi are surprisingly large, and are among the largest in the fungal kingdom,” says Dr. Maria Harrison, one of the principal investigators on the project. “The D. epigaea genome is 154 megabases (millions of base pairs), which is larger than that of the model plant Arabidopsis.”
D. epigaea is the third AM fungus to have its full genome sequenced. It is estimated to have diverged from the two previously sequenced species (Rhizophagus irregularis and Rhizophagus clarus) about 331 million years ago. That evolutionary distance is important, as it helps researchers to separate genomic characteristics unique to AM fungi from characteristics unique to Rhizophagus.
Fungi that form symbioses with roots secrete molecules that signal to nearby plants that they are ‘friendly’. In AM fungi these signaling molecules are large complexes of sugars and lipids (fats) called Myc factors. Myc factors are structurally similar to Nod factors – signaling molecules produced by rhizobia, another type of soil-dwelling symbiotic fungus; however, AM fungi lack enzymes similar to those used for Nod factor production. Therefore, biologists hypothesized that Myc factor synthesis must occur via a different pathway.
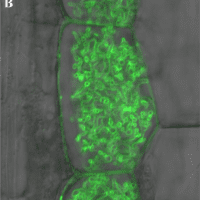
D. epigaea colonizes the interior of plant root cells, forming structures called arbuscules. The membrane at the plant-fungus interface and surrounding the plant cell is labeled with green fluorescent protein. A 10 micrometer bar in the lower left corner is provided for scale.
Image credit: Sun et al., 2018
In comparing the AM fungal genomes to genomes of free-living fungi, researchers identified a family of glycosyltransferase proteins (enzymes which add sugars to lipid molecules) that was significantly larger in AM fungi. Involvement of these proteins in Myc factor synthesis could explain the unusually large number of glycosyltransferases in AM fungi. If so, these findings would be the first to shed light on how Myc factors are produced.
Along with the genome of the fungus itself, researchers were also able to assemble the genomes of D. epigaea’s endosymbionts. “The most exciting result for me was the bacterial endosymbiont genomes,” says Dr. Xuepeng Sun, the lead author on the paper. “This was not expected at the beginning of the project, but we were happy to discover and assemble their genomes.”
While the genomes of bacterial endosymbionts have been sequenced before, this is the first time a mycorrhizal fungus and its endosymbionts have been sequenced together, which provides an opportunity to explore their co-evolution. The genomes of the endobacteria suggest that they are unable to synthesize the building blocks needed for nucleic acid (DNA and RNA) biosynthesis, and therefore must import them from the fungus. However, D. epigaea also requires these building blocks, which are costly to produce. Evidence from the D. epigaea genome suggests that the fungus has responded by expanding its ability to recycle nucleic acids, possibly to compete with its endosymbionts.
In addition to the knowledge gleaned from the D. epigaea and endosymbiont genomes, these sequences will be a valuable tool for future research on mycorrhizae, host-symbiont co-evolution, and fungal biology.
This research was supported by the National Science Foundation Plant Genome Research Program (grant no. IOS 1237993), the DOE Office of Biological and Environmental Research (grant no. DE-SC0012460), and the USDA Agriculture and Food Research Initiative (grant no. 2014-67013-21571).

