Interns 2019 BCBC Bioinformatics Course
About the Course
We are living in massive data times, and science is not an exception. New sequencing technologies are filling hard disks with terabytes of information, billion of sequences that need to be analyzed in a proper way. But not only sequence data is growing, gene expression and metabolite concentrations are analyzed by the hundreds or thousands, in a way that makes it difficult, if not impossible, to use familiar tools such as Excel. In this new world, bioinformatic skills are needed, not only by computational biologists, but by biologists and biochemists who find themselves analyzing many genes, proteins or metabolites at the same time.With this perspective, we aim to further bioinformatic skills within the postdoc community at Boyce Thompson Institute through this course. We try to keep it as simple as we can, with just one idea: “Show useful tools to resolve common problems found during the *omics data analysis”. For example, if I have two lists of hundred of genes, how can I combine them and find the common ones, or how can I analyze GO terms for my over-expressed genes, or how can download a chromosome region using Jbrowse or… there are dozens of examples.
Before you arrive — Online Bioinformatics Tools
A variety of tools exist online that may be used for bioinformatic analysis of small data sets. We cover these types of web tools in the slides below. Future lessons will focus on command line and programming methods of large data analyses. If you are overwhelmed by these slides, or the exercises therein, we will communicate an opportunity for a pre-workshop discussion.
Presenter: Suzy Strickler
Topics covered:
-
web based databases
-
web blast
-
genome browsers
-
sequence alignment
-
phylogeny
-
primer design
Estimated Time
- Slides and exercises: 1:00 h
Materials
BEFORE 06/12/19 — Setting Up the BCBC Course Virtual Machine
Steps:
1. Download and install VirtualBox following the package instructions, as well as the VirtualBox Extension Pack. For more info consult the user manual. Please download and install the latest VirtualBox version for your operating system.
2. Check if you have a 64-bit or 32-bit system (see these links for instructions: Windows or MacOS) and download the BCBCBIC2019_debian.ova file from below. You can also click the link to download it. If you have a 32-bit system, please send us an email. Some Windows 64 bits systems don’t activate the acceleration VT-x/AMD-V, which prevents running the virtual machine. You will need to enable the VT-x/AMD-V in the Bios of your computer.If you are running a 32-bit computer you can come to our office to find an alternative 64-bit laptop that you may use for the course.
3. Create a VM Folder in your system and copy/move the .ova file.
4. Open the VirtualBox program.
6. Select the option File > Import Appliance![]()
7. Click “Open Appliance”. Select the .ova file and click “Continue”.
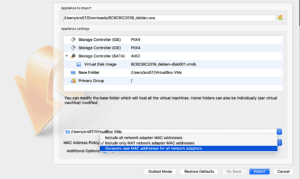
8. Enable “Generate new MAC addresses for all network adapters” and click “Import”.
9. You must attend bioinformatics hour Tuesday 1 – 2 pm or Wednesday 1 – 2 pm in the BTI Resource Center to have your VM installation confirmed.
Troubleshooting
- I have enabled virtualization but VirtualBox still gives an error asking to “Enable vt-X” or similar.
Make sure you have disabled hyper-v. This can be done by following these instructions.
- I have downloaded both the VirtualBox software and the virtual machine file and enabled virtualization, but it still will not run.
Make sure you have a 64-bit machine and have followed the above steps precisely, especially enabling . Come to our office or email us for further troubleshooting if you cannot find the problem.
06/13/19 — UNIX Command-Line Intro, Part 1
Presenter: Suzy Strickler
Topics covered:
- Terminal file system navigation
- Wildcards, shortcuts and special characters
- File permissions
- Compression UNIX commands
- Networking UNIX commands
Estimated Time
- Lecture and exercises: 2:00 h
Materials
06/20/19 — UNIX Command-Line Intro, Part 2
Presenter: Adrian Powell
Topics covered:
- Basic NGS file formats
- Text files manipulation commands
- Command-line pipelines
- Introduction to bash scripts
Estimated Time
- Lecture and exercises: 2:00 h
Materials
06/27/19 — Next Gen Sequencing
07/02/19 — Introduction to R & Differential Expression, Part 1
Presenter: Adrian Powell
Prerequisites
- Make sure that you have “gene_count_matrix.csv” file in the “Slch04_demo” directory in the Desktop directory of your VM. Please let us know before class if you missed a previous session, or were unable to complete the exercises, and do not have the necessary files.
Topics covered
- Brief introduction to R
- Data types
- Differential Expression
Estimated Time
- Lecture and examples: 2:00 h
Materials
07/11/19 — Introduction to R & Differential Expression, Part 2
Presenter: Adrian Powell
Topics covered
- General pipeline for differential expression analysis with an emphasis on edgeR
- Data exploration
- R graphs
Estimated Time
- Lecture and examples: 2:00 h
Materials