News
International team collaborates to decipher the Asian citrus psyllid genome
The Asian citrus psyllid (ACP) transmits CLas by feeding on citrus leaves. New genomic insights may help to break the cycle. Credit: Heck Lab
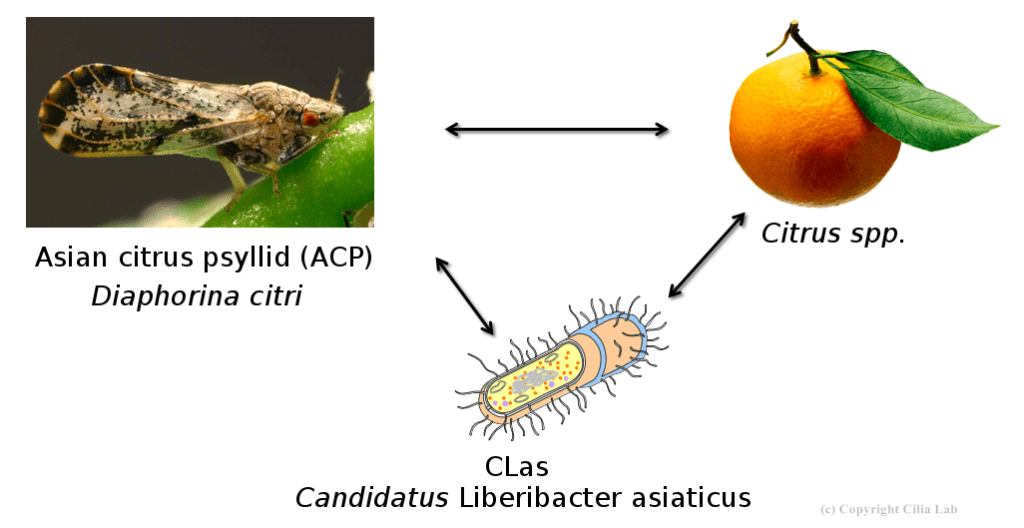
The Asian citrus psyllid (D. citri, or ACP) is the insect vector that spreads the bacterium responsible for citrus greening disease (Candidatus Liberibacter asiaticus, or CLas). While much research attention has been given to CLas and the citrus plants it infects, the insects have received less attention. Thanks to new research at the Boyce Thompson Institute (BTI) and partner institutions, attacking the ACPs may become the newest solution to ending citrus greening for good.
BTI’s Mueller and Heck Labs, in collaboration with 21 partner institutions, recently published a draft assembly and annotation of the D. citri genome comprising 530 manually curated gene models and 20,000 automatically predicted genes. Researchers published their community annotation in Database.
“It is essential to have detailed genetic and genomic knowledge to tackle the psyllid, but a lack of this information had been very problematic,” according to Surya Saha, BTI senior bioinformatics analyst and project leader. “Now, an annotated ACP genome exists, containing a wealth of information needed to develop molecular-based solutions to combatting this pest.”
The detailed knowledge lies in the DNA of the ACP – or its genome sequence. Using multiple different technologies, the scientists determined the sequence of the ACP genome. Next, they were faced with the difficult challenge of interpreting that sequence. “The genome of the ACP contains thousands of genes. Some of them are similar to other insects, and we can infer their complete sequence and function by comparing the ACP sequence to sequences from other organisms. Others are totally different, and specific to the ACP. These are much more difficult to study and critically important” according to Michelle Heck, BTI assistant professor and project leader.

Credit: USDA
Genes in both categories have to be manually corrected in many cases, to produce the most complete and accurate information from the data. The group produced a new gene set, which combines automatic gene predictions and manual curations. “It is truly a team effort” says Lukas Mueller, BTI assistant professor and project leader. The manual curation effort focused on genes that the group think are important for transmission of the citrus greening bacterium.
“The automated genome is similar to scrambled words: our team still had to find the important components and put them in an order that is understandable,” according to Prashant Hosmani, a BTI bioinformatics analyst and co-first author. “By manually annotating these specific gene families, we’ve created a more comprehensive blueprint that can be used to target ACP genes with genome editing technologies such as RNAi and CRISPR.”
In addition, the study is also a demonstration of how to successfully combine various levels of expertise and protocols to harness the crowd to address an important research question. The combined efforts of a global consortia has grown to become a powerful strategy for research innovation. With 36 curators involved in the project, the development of project infrastructure, workflows, trainings, and document sharing platforms was critical to the project’s success.
“The workplace of tomorrow will involve large collaborations and plenty of remote communication,” according to Saha. “This project exemplifies how successful large consortium projects can succeed, given the right tools and guidance. We are writing up the procedures we followed and optimized as a reference for others in the community to implement in their projects”
“What makes the project especially unique is the combination of expert curators and students working together,” said Mirella Flores Gonzales, a BTI database programmer and co-author. “The project included undergraduate and graduate students from community colleges and large research universities alike. Not only were students learning from experience, but they now are equipped to train the next cohort of project partners.”
“The student co-authors include 2 generations of BTI PGRP summer interns who were co-mentored by scientists in the Heck and Mueller labs.“ Saha added.
The community annotation process will be continued by future cohorts of student annotators and scientists to improve the next version of the genome and annotation. The official annotation and additional information on the project can be found at citrusgreening.org. More information about the ACP genome and curation projects can be found here. Click here for the link to the annotation.
All open-access fees, student annotators and post-docs were funded through USDA-NIFA grant 2015-70016-23028. The genome sequencing was supported by grants from the NIH National Center for Research Resources (5P20RR016469), USDA-ARS, Ft. Pierce, FL and a grant from the Citrus Research Board, Inc. (217 North Encina, PO Box 230, Visalia, CA 93279).


